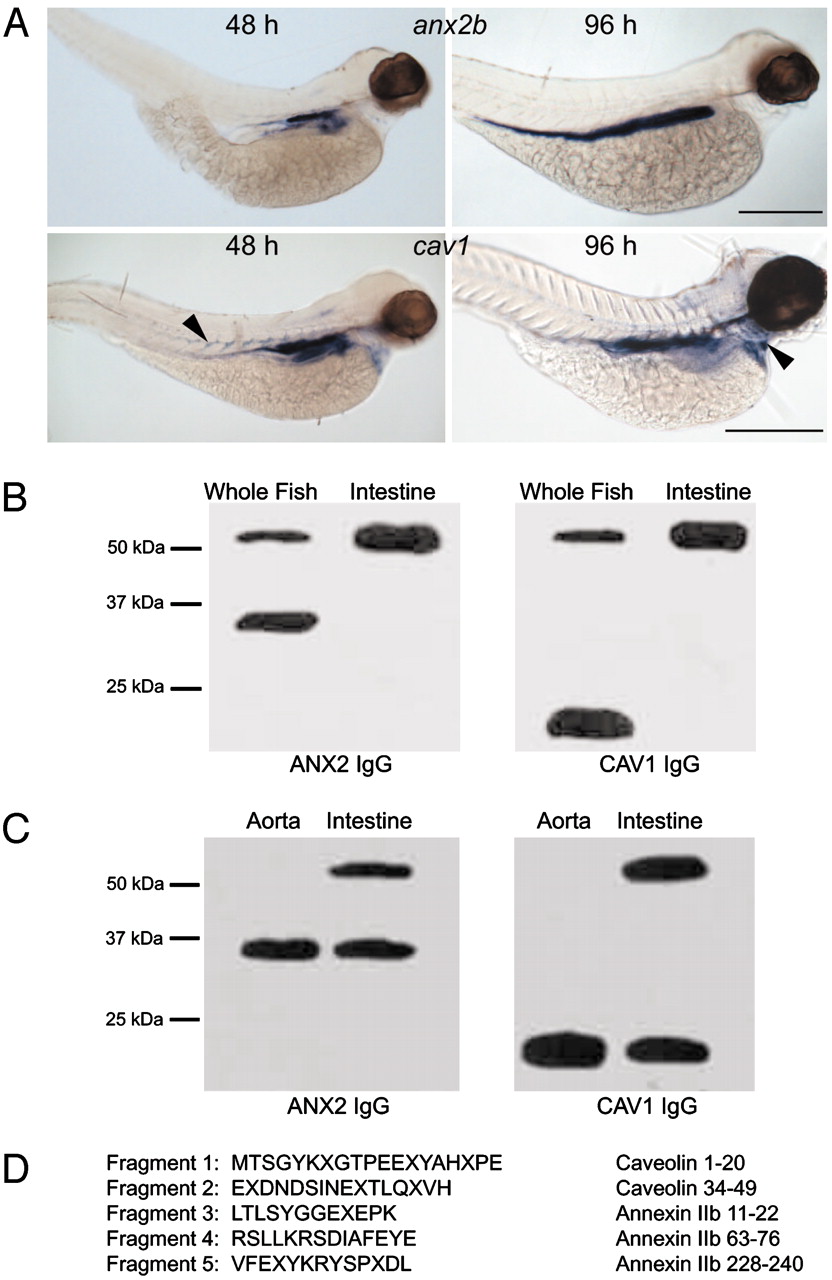
Fig. 1 CAV1 and ANX2b can form a stable heterocomplex. (A) Expression of cav1 and anx2b in zebrafish larvae. Embryos were fixed in 4% paraformaldehyde and probed with digoxigenin-labeled antisense RNA as described in ref. 19. (Upper) Lateral views of embryos probed for anx2b at 48 hpf (Left) and 96 hpf (Right). Note strong expression in the intestinal epithelium. (Scale bar, 500 μm.) (Lower) Lateral views of embryos probed for cav1 at 48 hpf (Left) and 96 hpf (Right). Expression is concentrated in the intestinal epithelium, but cav1 can also be seen in the somite boundaries at 48 hpf (Left, arrowhead) and in the heart ventricle at 96 hpf (Right, arrowhead). (Scale bar, 500 μm.) (B) Identification of a CAV1–ANX2b heterocomplex. Equal amounts of protein (20 μg) isolated from adult fish or adult fish intestine were resolved by SDS/PAGE and immunoblotted with ANX2 IgG or CAV1 IgG. The data are representative of five independent experiments. (C) Equal amounts of protein (20 μg) isolated from the aorta or intestine of C57BL/6 mice were resolved by SDS/PAGE and immunoblotted with ANX2 IgG or CAV1 IgG. The data are representative of three independent experiments. (D) The ≈55-kDa band was subjected to IP from adult intestine by using CAV1 IgG as described in ref. 11 and resolved by SDS/PAGE. The 55-kDa band was recovered from the gel and digested with trypsin; the resulting fragments were resolved by SDS/PAGE and transferred to nylon membrane. Five of the fragments were sequenced by mass spectrometry. The sequence of each fragment is shown along with the region to which they correspond in CAV1 or ANX2b. The letter "X" signifies an unidentified amino acid residue.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Proc. Natl. Acad. Sci. USA