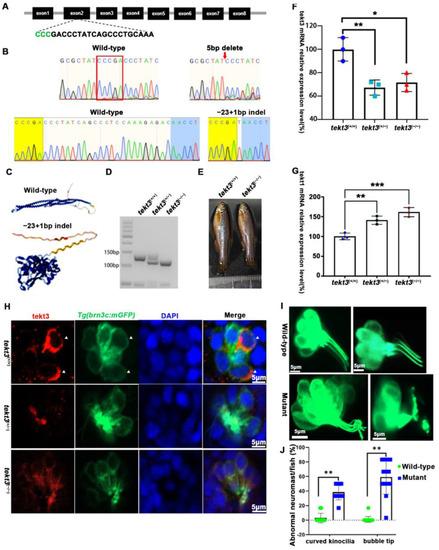
tekt3 mutant exhibited only neuromast HC kinocilium defect. (A) The CRISPR/Cas9-mediated genome editing targeted the exon 2 of tekt3 locus; the gRNA core sequence is shown. (B) Two tekt3 mutant alleles were verified by sequencing. (C) The (Alphfold2.0) predicted protein structure of one mutated Tekt3 is shown side by side with a normal protein. (D) A typical genotyping result of tekt3 mutant. (E) A 6 mpf homozygous tekt3 mutant looked normal. (F) Significant reduction in tekt3 mRNA level can be seen in both homozygous and heterozygous tekt3 mutants. (G) The tekt1 mRNA level in 5 dpf fish were significantly up in both homozygous and heterozygous tekt3 mutants. (H) Tekt3 was decreased in neuromast HCs of both homozygous and heterozygous tekt3 mutants (5 dpf). The arrowheads are pointed to tekt3 positive cells. (I) The abnormal neuromast HC hair bundle or kinocilia of the 5 dpf tekt3 mutant. (J) Almost all mutant neuromasts (n = 78) showed abnormal kinoclium tips while wild-type (n = 112) rarely possessed the defect. * p < 0.05; ** p < 0.01; and *** p < 0.001.
|