Fig. 3
- ID
- ZDB-FIG-250414-3
- Publication
- Ye et al., 2025 - Zebrafish as a model for investigating Klebsiella pneumoniae-driven lung injury and therapeutic targets
- Other Figures
- All Figure Page
- Back to All Figure Page
|
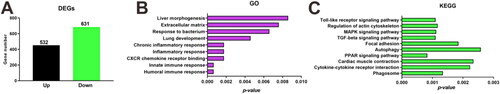
Key signaling pathways revealed by transcriptomic analysis of Klebsiella pneumoniae-induced lung injury in zebrafish. (A) Changes in the number of differentially expressed genes (DEGs). Genes with significant upregulation and downregulation were identified using edgeR analysis (FDR < 0.05). (B) GO enrichment analysis highlighted key biological processes, including liver morphogenesis, extracellular matrix remodeling, response to bacterial infection, lung development, chronic inflammation, and CXCR cytokine receptor binding. Fisher’s exact test was used for enrichment analysis, with p < 0.05 considered significant. (C) KEGG enrichment analysis identified significant enrichment in several key signaling pathways, including the Toll-like receptor pathway, MAPK signaling pathway, TGF-β signaling pathway, PPAR signaling pathway, cytoskeletal regulation, and phagosome pathway. Statistical analysis was performed using Fisher’s exact test (p < 0.05). |