Fig. 2
- ID
- ZDB-FIG-240916-178
- Publication
- Cömert et al., 2024 - HSP60 chaperone deficiency disrupts the mitochondrial matrix proteome and dysregulates cholesterol synthesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
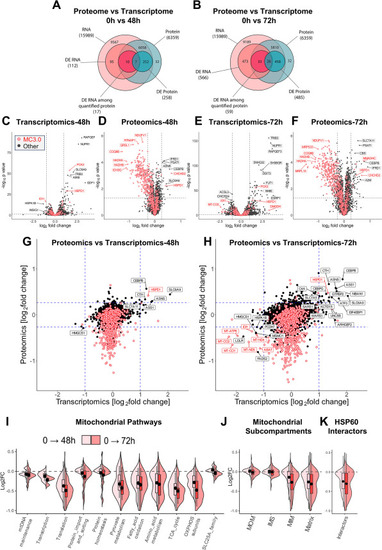
HSP60 dysfunction severely regulates the mitochondrial proteome, but not the transcriptome. (A–B) Venn diagrams representing the number of quantified and differentially expressed genes in transcriptomics and proteomics. D423A cells at 0 h were compared to 48 h (A) or 72 h induced D423A cells (B). The pairing of quantified genes of transcriptomics and proteomics was performed through Ensembl gene IDs. (C-F) Volcano plots showing the comparison of induced D423A cells at 48 h and 72 h with 0 h for quantified transcriptome (C, E), and proteome (D, F). Cut-off values are indicated with dashed lines. As indicated in Figure 2 C, Mitochondrial proteins based on MitoCarta3.0 (MC3.0; [ 2 ]) are highlighted with red points. (G, H) Comparison plot of log2fold changes of Proteomics versus Transcriptomics at 48 h (G) and 72 h (H). Cut-off values are indicated with dashed lines. Mitochondrial proteins are highlighted in red. Genes significantly up- or down-regulated at both transcript and protein levels are labeled. (I-K) Violin plots distinguishing proteins from major mitochondrial pathways (I), mitochondrial sub-compartments (J), and HSP60 interactors (K), Induced D423A cells at 0 h in comparison to 48 h (light red) or 72 h induced D423A cells (dark red), respectively. The violin plots reflect all quantitated proteins in the respective pathways, compartments, and groups; violin widths are kept constant in the plots. Cut off values for transcriptomics: Benjamini-Hochberg adjusted p-value <0.05 and |log2FC|>1. Cut off values for proteomics: p-value <0.05 and |log2FC|>0.26 Abbreviations: DE, differentially expressed; FC, fold change; IMS, intermembrane space; MIM, mitochondrial inner membrane; MOM, mitochondrial outer membrane (See also the legend of Figure 1 ). |