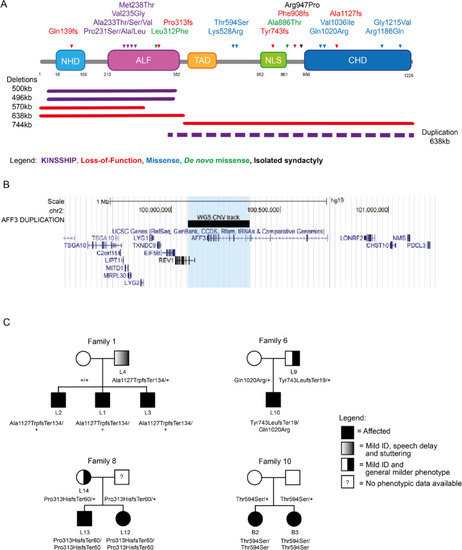
AFF3 variants. A Schematic protein structure of AFF3 (NM_002285.3) with its N-terminal homology domain (NHD, cyan), the AF4-LAF4-FMR2 (ALF, pink) domain [2, 3, 5] containing the degron motif, a Serine-rich transactivation domain (TAD, yellow) [6], a bipartite nuclear/nucleolar localization sequence (NLS, green), and the C-terminal homology domain (CHD, blue) [4] showing positioning of all AFF3 coding variants mentioned in the text. Missense and truncating variants are shown above, while extents of microdeletions and microduplications are depicted below the structure with continuous and dashed lines, respectively. The variants are color coded: loss-of-function (truncation and deletion) in red, biallelic missense outside the degron in blue and KINSSHIP-associated missense variants, deletion, and duplication in purple. The de novo missense identified in individuals M1 and M2 are shown in green. While the p.(Arg947Pro) shown in black was shown to segregate with isolated syndactyly [12], we have also identified it in individuals with no digit abnormalities. The 500 kb KINSSHIP deletion was originally wrongly defined as encompassing the entirety of the gene [13] but was later shown to only remove exons 4 to 13 of AFF3 [11, 14, 15]. B UCSC genome browser snapshot of the Chr2 99.5 to 101.4-Mb region showing the genes mapping to this interval. The extent of the duplication identified in the DUP1 individual that encompasses exon 10 to exon 24 of AFF3 and exon 1 to 3 of REV1 is indicated by the black bar and the light blue shadow. C Examples of pedigrees of transmitting affected families suggesting semi-dominance
|