|
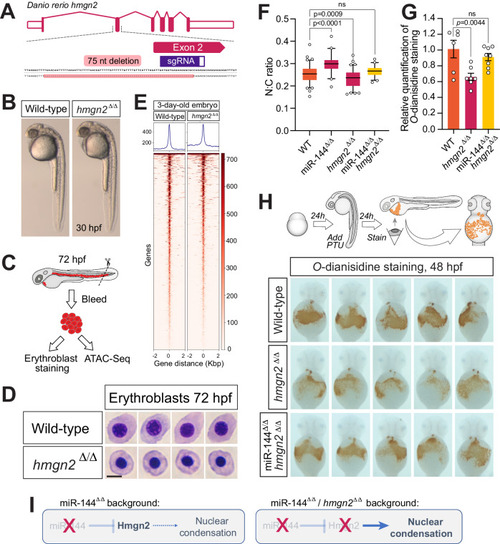
Reduction of Hmgn2 activity rescues the loss of miR-144 in erythropoiesis. A Schematic representation of hmgn2 gene from Danio rerio and CRISPR/Cas9 induced deletion. B Maternal-zygotic Hmgn2 (hmgn2Δ/Δ) mutants at 30-hpf. C Cartoon describing the collection of peripheral blood from embryos at 72-hpf for ATAC-Seq and cellular analysis. Cartoon is adapted with permission from ref. 17. D May–Grünwald–Giemsa staining of peripheral blood cells isolated from hmgn2Δ/Δ and wild-type siblings at 72-hpf. Scale bar indicates 5 µm in length. Data representative of two independent bleeding and staining experiments. E Heatmaps display variations in chromatin accessibility, determined through ATAC-Seq analysis of three separate samples obtained from erythrocytes isolated from peripheral blood at 72 h post-fertilization (hpf) in both hmgn2Δ/Δ and wild-type sibling embryos. F Quantitative analysis of the nucleocytoplasmic ratio of erythrocytes isolated from miR-144Δ/Δ, miR-144Δ/Δ/hmgn2Δ/Δ and wild-type siblings stained with May–Grünwald–Giemsa to rescue miR-144Δ/Δ phenotype. Individual cells (n = 89 cells from wild-type, n = 41 from miR-144Δ/Δ, n = 85 from hmgn2Δ/Δ, and n = 45 from 144Δ/Δ/hmgn2Δ/Δ) from pools of bled embryos are analyzed in each case. Wild-type and hmgn2Δ/Δ data are derived from two independent pools of bled embryos. P values from one-way ANOVA with Dunnett’s multiple comparisons test. Boxes enclose data between the 25th and 75th percentile, with a horizontal bar indicating the median. Whiskers enclose 5th to 95th percentiles. G Relative quantification of the area stained with O-dianisidine in the embryos shown in (H). Data represent mean ± standard error of the mean. P values from one-way ANOVA with Dunnett’s multiple comparisons test. HO-dianisidine staining of 2-dpf embryos to reveal hemoglobinized cells. Embryos are pre-treated with PTU to induce mild oxidative stress to exacerbate any defect in erythropoiesis. n = 6 of wild-type, n = 7 of hmgn2Δ/Δ, and n = 8 of miR-144Δ/Δ/hmgn2Δ/Δ individual embryos are analyzed. I Schematic of genetic probing of miR-144/hmgn2 regulatory axis. In miR-144 mutant embryos, the absence of miR-144-mediated repression drives Hmgn2 overexpression, which in turn leads to reduced chromatin compaction. In the double miR-144Δ/Δ/hmgn2Δ/Δ embryos, the absence of Hmgn2 activity rescues the effect of the loss of miR-144 in nuclear condensation.
|