Fig. 2
- ID
- ZDB-FIG-240315-29
- Publication
- Walter et al., 2024 - Functionally-instructed modifiers of response to ATR inhibition in experimental glioma
- Other Figures
- All Figure Page
- Back to All Figure Page
|
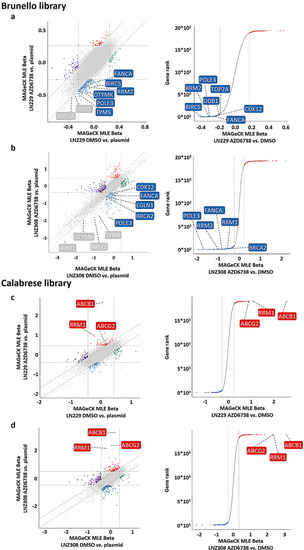
CRISPR-Cas9 genome-wide knockout and activation screens identify novel combination partners for ATRi. |