Fig. 4.
- ID
- ZDB-FIG-230228-86
- Publication
- Heng et al., 2023 - Cpeb1b-mediated cytoplasmic polyadenylation of shha mRNA modulates zebrafish definitive hematopoiesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
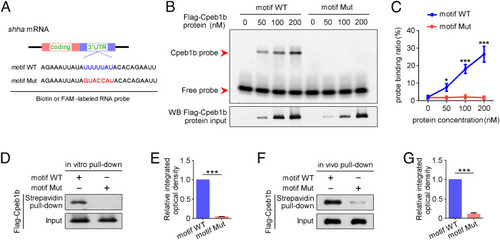
The CPEB motif in shha mRNA is important for Cpeb1b binding. (A) Diagram of biotin or FAM-labeled CPEB motif probe. The WT CPEB binding motif is in blue color, and the mutant CPEB binding motif is in red color. The sequence of motif WT and motif Mut probes is listed. (B) EMSA measuring the Cpeb1b binding with FAM-labeled motif WT and motif Mut probes. The protein concentration ranged from 0 nM to 200 nM. (C) Quantification of each band by gray analysis (Gel-Pro analyzer). The probe binding ratio at each group was determined by (RNA-protein)/[(free RNA) + (RNA-protein)]. GraphPad Prism was used for statistical analysis with Student’s t test. Statistic data were shown as mean ± SD. The P value was used for significance evaluation, *P < 0.05, ***P < 0.001. (D) WB showing the purified Flag-Cpeb1b protein pulled down with biotin-labeled CPEB motif probe. (E) Quantification of relative Flag-Cpeb1b protein level using gray analysis. Error bar, mean ± SD. The P value was calculated by Student’s t test, ***P < 0.001. (F) WB showing the Flag-Cpeb1b protein from Flag-cpeb1b mRNA-injected embryo extracts pulled down with biotin-labeled CPEB motif probe. (G) Quantification of relative Flag-Cpeb1b protein level using gray analysis. Error bar, mean ± SD. The P value was calculated by Student’s t test, ***P < 0.001. |