FIGURE 3
- ID
- ZDB-FIG-220722-19
- Publication
- Mukaigasa et al., 2021 - The developmental hourglass model is applicable to the spinal cord based on single-cell transcriptomes and non-conserved cis-regulatory elements
- Other Figures
- All Figure Page
- Back to All Figure Page
|
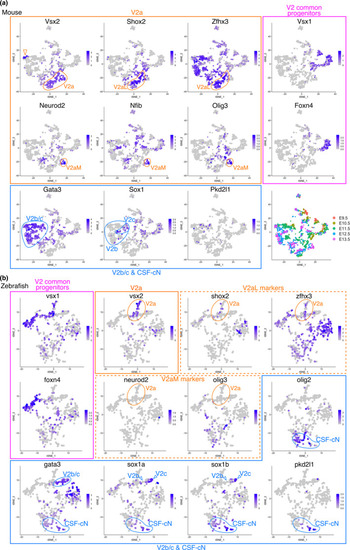
Distinct gene expression profiles in V2 INs between mice and zebrafish. tSNE plot showing the cells with V2 IN identity derived from mouse (a) and zebrafish (b) embryonic spinal cords. The expression levels of the indicated genes were visualized on the tSNE plot. Plots for V2 common progenitor markers are enclosed in magenta boxes. Plots for V2a IN markers are enclosed in yellow boxes. Plots for the V2b/c IN and CSF‐cN markers are enclosed in blue boxes. The bottom right panel in (a) shows the embryonic day when the cells are corrected. Within the tSNE plots, specific subpopulations (V2a, V2aM, V2aL, V2b, V2c, and CSF‐cNs) are indicated. The expression profiles of V2aL and V2aM markers (shox2, zfhx3, neurod2, and olig3) in zebrafish were different from those in mice (yellow broken‐line boxes in b). The Vsx2‐high minor population was found in the mouse tSNE plot as a distinct cluster (open arrowhead in a), which is likely to be type‐I V2a neurons, and other major V2a INs are probably type‐II neurons (Hayashi et al., |