Fig. 3
- ID
- ZDB-FIG-220701-68
- Publication
- Sato et al., 2022 - Identification of embryonic RNA granules that act as sites of mRNA translation after changing their physical properties
- Other Figures
- All Figure Page
- Back to All Figure Page
|
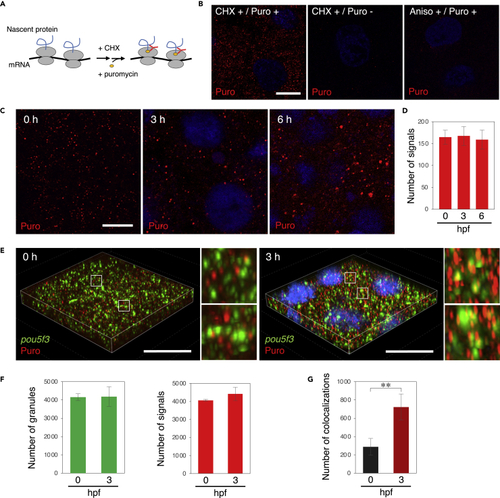
Embryos start to translate pou5f3 mRNAs at the sites of granules during the mitotic cleavage stage (A) A schematic view for visualization of newly synthesized peptides by ribopuromycylation. After pretreatment with CHX, embryos were treated with CHX and puromycin, which was detected by anti-puromycin antibody after washing out free puromycin. Note that this method also enables detection of puromycylated peptides released from polysomes. (B) Detection of newly synthesized peptides (red) with anti-puromycin antibody in cleavage-stage embryos stimulated with (+) or without (−) CHX and puromycin (Puro). As a control, embryos were pretreated with anisomycin (Aniso) instead of CHX. DNA is shown in blue. (C) Detection of newly synthesized peptides (red) in embryos at 0, 3, and 6 h post fertilization. DNA is shown in blue. (D) Numbers of signals of newly synthesized peptides per 3,600 μm2 in embryos at 0, 3, and 6 hpf were counted (means ± standard deviations; n = 3). Similar results were obtained from three independent experiments. (E) 3D images of SIM for pou5f3 mRNA (green) and newly synthesized peptides (red) in embryos at 0 and 3 h post fertilization. DNA is shown in blue. Insets show enlarged views of the boxed regions. Bars, (B and C) 10 μm, (E) 20 μm. (F) Numbers of pou5f3 RNA granules (left) and newly synthesized peptides (right) per 28,800 μm3 in embryos at 0 and 3 hpf were counted (means ± standard deviations; n = 3). (G) Numbers of colocalizations of pou5f3 RNA granules and newly synthesized peptides per 28,800 μm3 in embryos at 0 and 3 hpf were counted (means ± standard deviations; n = 3). Similar results were obtained from two independent experiments. ∗∗p < 0.01 (Student’s t test). |