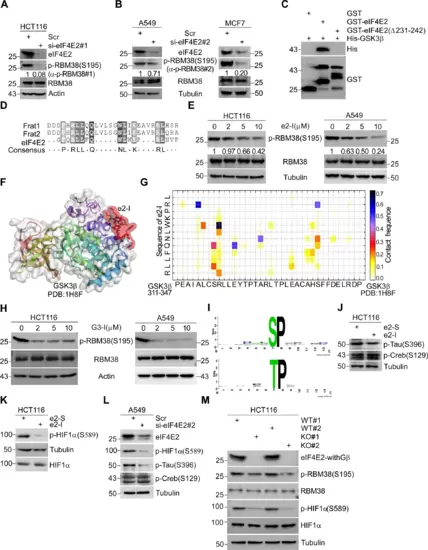
Fig. 1: eIF4E2 regulates GSK3β proline-directed kinase activity. A, B Depletion of eIF4E2 inhibits the phosphorylation of RBM38-Ser195. eIF4E2 siRNA#1 was transfected into HCT116 cells (A) or siRNA#2 was transfected into A549 or MCF7 cells (B) for 72 h, followed by WB with indicated antibodies. Antibody p-RBM38#1 or p-RBM38#2 was used to detect p-RBM38(Ser195) as indicated. C Identifying the GSK3β binding motif of eIF4E2 by GST pull-down assay. GSK3β directly interacts with eIF4E2, but not with eIF4E2 mutant lacking amino acid from 231 to 242 (Δ231–242). D The GSK3β binding motif of eIF4E2 or FRAT (Frat1 and Frat2) were aligned. E e2-I inhibits the phosphorylation of RBM38-Ser195. Cells were treated with different concentrations of e2-I (2, 5, 10 μM) or scrambled e2-S (10 μM) for 24 h, followed by WB. F CABS-dock showed the binding of e2-I to GSK3β. The crystallographic structure of GSK3β (PDB ID: 1H8F) was used and the interaction interface was highlighted in red. G The contact diagram of GSK3β with e2-I, proposed by CABS-dock web server. H G3-I inhibits the phosphorylation of RBM38-Ser195. I The most over-represented motif is proline-directed serine/threonine regulated by eIF4E2-GSK3β pathway. Phosphorylation motifs were extracted by using Motif-x algorithm and threshold for significance was set to P < 0.000001. J, K e2-I inhibits the proline-directed phosphorylation, including Tau-Ser396 (J) and HIF1α-Ser589 (K) but does not affect the phosphorylation of Creb-Ser129 that is a serine site within priming motif. L Depletion of eIF4E2 inhibits the Tau-Ser396 and HIF1α-Ser589. eIF4E2 siRNA#2 was transfected into A549 cells for 72 h, followed by WB with indicated antibodies. M Knockout of eIF4E2 isoforms with GSK3β binding motif (eIF4E2-withGβ) inhibits the RBM38-Ser195 and HIF1α-Ser589. Lysis of eIF4E2-KO HCT116 (KO) and isogenic wild-type HCT116 (WT) cells, followed by WB with indicated antibodies.
|