Fig. 7
- ID
- ZDB-FIG-220304-30
- Publication
- Forcella et al., 2021 - Characterization of three sialidases from Danio rerio
- Other Figures
- All Figure Page
- Back to All Figure Page
|
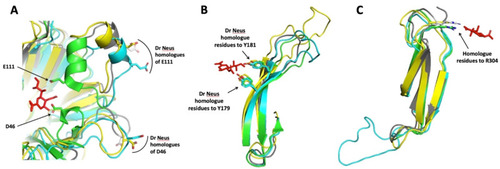
Superimposition of portions of NEU2/DANA complex crystal structure (PDB ID 1VCU) with Dr Neus predicted structures. (A) Overlapping of Hs NEU2 portion bearing the D46 and E111 residues with Dr Neu3.1, 2 and 3. The competitive inhibitor DANA interacts with the side chains of these acidic residues, located in α-helix portions of two mobile loops [13]. The homologue residues of Dr Neus 3D models are located in loops far away from the active site crevice. An α-helix portion is present only in the loop containing E111, as expected by the sugar-induced form of Hs NEU2 (PBD ID 1SO7) used as a template for modelling. (B) DANA interacts with the hydroxyl groups of tyrosine 179 and 181 and the aromatic residues, located in beta sheets B (174–182) and C (191–199) of the third blade of the propeller [13]. The homologue residue positions are highly conserved in Dr Neus. Note the variable and increasing length of the loop connecting the two antiparallel beta sheets. (C) DANA interacts with the side chain terminal portion of the arginine 304 [13] and the homologue residues of Dr Neus are located in topologically equivalent positions. Note the difference in the length of the loop connecting beta sheets A (275–280) and B (291–298) of the fifth blade of the propeller in Dr Neu3.1 compared to the other sialidases. The amino acid numbers are referred to Hs NEU2 [13]. Color code: DANA red, Hs NEU2 green; Dr Neu3.1 light blue; Dr Neu3.2 grey and Dr Neu3.3 yellow. |