Fig. 1
- ID
- ZDB-FIG-210507-8
- Publication
- Sommer et al., 2021 - Disruption of Cxcr3 chemotactic signaling alters lysosomal function and renders macrophages more microbicidal
- Other Figures
- All Figure Page
- Back to All Figure Page
|
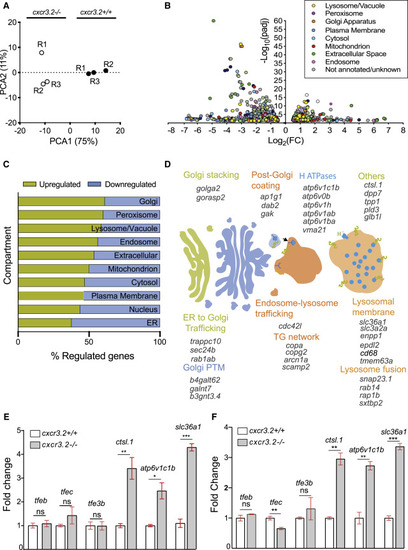
Disruption of Cxcr3.2 signaling transcriptionally induces genes related to lysosomal function and intracellular vesicle trafficking (A) Principal-component analysis (PCA) of cxcr3.2 mutant (cxcr3.2−/−) and WT (cxcr3.2+/+) transcriptomes. PCA analysis was performed in R on variance-stabilizing transformed (vst) data, using the DESeq2 plotPCA command. (B) Volcano plot of cxcr3.2 mutant versus WT differentially expressed genes. Genes are classified and color-coded by cellular compartment annotation. Compartment annotations were obtained from http://geneontology.org according to the GO cellular component and from KEGG pathways. (C) Distribution of upregulated (yellow) and downregulated (blue) genes, classified by compartment as above. Lysosomal, Golgi, and peroxisome-related genes are more commonly upregulated in cxcr3.2 mutant macrophages. (D) Graphical representation of induced genes exerting key functions in Golgi and lysosomal pathways. ER, endoplasmic reticulum; PTM, post-translational modification; TG, trans-Golgi. (E and F) Expression fold change of representative lysosomal markers and transcriptional regulators of lysosomal functions of cxcr3.2 mutant and WT FACS macrophages, as determined by qPCR (E) or RNA-seq analysis (F). qPCR analysis confirmed that overall lysosomal function is increased in cxcr3.2 mutants as indicated by the upregulation of lysosomal function markers ctsl.1, atp6v1c1b, and slc36a1, whereas the expression of the lysosomal biogenesis regulators tfeb, tfe3, and tfec remained unaltered. Three biological samples of 150–200 larvae were used, and three technical replicates were conducted. Data were analyzed using a two-tailed t test and results are shown as mean ± SEM (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; ns, not significant [p > 0.05]). |