Fig. 2
- ID
- ZDB-FIG-210128-183
- Publication
- Takita et al., 2020 - eys+/- ; lrp5+/- Zebrafish Reveals Lrp5 Can Be the Receptor of Retinol in the Visual Cycle
- Other Figures
- All Figure Page
- Back to All Figure Page
|
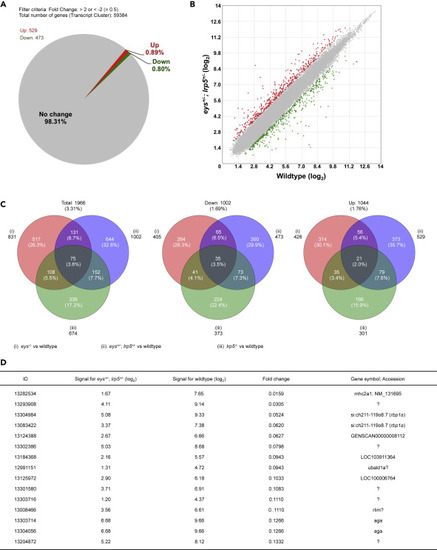
Figure 2. Global Gene Expression Analysis of the Digenic eys+/-; lrp5+/− Zebrafish Eyes Identifies the retinol binding protein 1a (rbp1a) Gene. (A and B) About 1.69% (1,002 genes) were 2-fold changed among the 59,384 transcript clusters (genes) in the eys+/-; lrp5+/− zebrafish eyes compared with the eyes of wild-type siblings (A). About 0.80% (473 genes) of the total were 2-fold downregulated and 0.89% (529 genes) were 2-fold upregulated (B, green and red spots, respectively). (C) About 3.31% (1,966 genes) were 2-fold changed, 1.69% (1,002) genes were 2-fold downregulated, and 1.76% (1,044 genes) were 2-fold upregulated in the eys−/−, lrp5−/−, or eys+/-; lrp5+/− eyes compared with the eyes of wild-type siblings (left, middle, and right panels, respectively). Among 0.80% (473 genes) 2-fold downregulated in the eys+/-; lrp5+/− eyes (middle panel), 21.1% (100 genes) were overlapped with the eyes of eys−/− siblings and 14.6% (108 genes) were overlapped with the eyes of lrp5−/− siblings. (D) Genes whose expression levels were dramatically changed were extracted. “Fold change” in the fourth column indicates the ratio (the bare value of the signal for eys+/-; lrp5+/− divided by that for the wild-type). The ratio for si:ch211-119o8.7 (the rbp1a gene, third and fourth rows) was identified as the gene remarkably downregulated in the eys+/-; lrp5+/− eyes (fold changes, 0.0524 (ID: 13304984) and 0.0620 (13083422)). “Signal for eys+/-; lrp5+/−” (second column) and “Signal for wild type” (third column) are shown in (log2). |