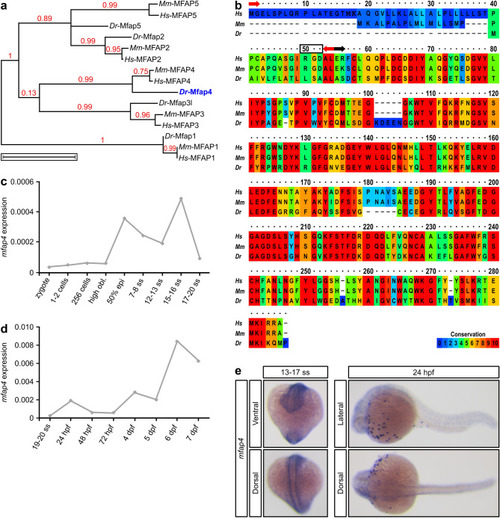
Alignment of vertebrate Mfap4 proteins and mfap4 expression during zebrafish development. (a) Phylogenetic analysis of human (Homo sapiens, Hs) MFAP family members and their murine (Mus musculus, Mm) and piscine (Danio rerio, Dr) orthologs. Scale bar represents 1 amino acid substitution per site. (b) Amino acid sequence alignment of human, murine and zebrafish MFAP4 proteins displayed in ClustalX color scheme. Red arrows demarcate the start and end of the 16 amino acid signal peptide (www.uniprot.org); black box highlights the RGD sequence; black arrow indicates the start of the fibrinogen-related domain (FReD). (c–e) Zebrafish mfap4 expression analysis by (c,d) quantitative RT-PCR (mean expression in twenty pooled embryos/larvae per time point) relative to the expression of the housekeeping gene actin, beta 1 (actb1) and by (e) whole-mount RNA in situ hybridization (purple staining)—ventral (13–17 somite stage, ss), dorsal (15–17 ss and 24 h post fertilization, hpf) and lateral (24 hpf) views. Phylogenetic analysis in panel (a) was performed with Phylogeny.fr online software (available at www.phylogeny.fr)63,64. Alignment in panel (b) was performed with the online PRALINE Multiple Sequence Alignment software (Vrije Universiteit Amsterdam, Centre for Integrative Bioinformatics VU, available at www.ibi.vu.nl/programs/pralinewww/). Graphs in panels (c) and (d) were generated with Microsoft Excel (for Mac 2011, version 14.7.7). This figure was created with Inkscape software version 0.92 (available at https://www.inkscape.org/).
|

