Fig. 6
- ID
- ZDB-FIG-200323-16
- Publication
- Hoshijima et al., 2019 - Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
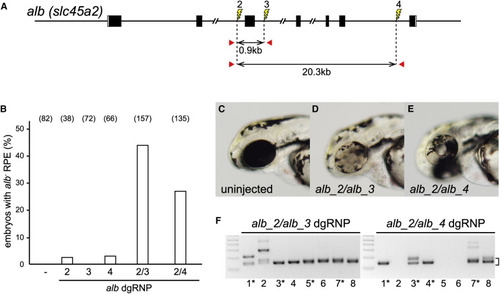
Deletion Mutations Are Readily Generated by Simultaneously Targeting Two Sites on a Chromosome (A) dgRNPs were designed to target sites (lightning bolts) in intron 2 (alb_2), intron 3 (alb_3), or intron 6 (alb_4). The approximate distances between target sites and the locations of primer pairs (red arrows) used to detect deletion mutations that result in joining of target sites are indicated. (B) Uninjected embryos (−) or embryos injected with individual or pairs of dgRNPs targeting alb locus introns were inspected for pigmentless patches of tissue in the RPE at 2–3 dpf. The incidence of embryos with alb mutant tissue is plotted (numbers of scored embryos are indicated). (C–E) Embryos injected with dgRNP pair alb_2 and alb_3 (D) or alb_2 and alb_4 (E) often exhibited large clones of pigmentless mutant tissue not seen in the eyes of uninjected embryos (C). (F) Deletion mutations were detected in the genomes of F0 embryos injected with pairs of dgRNPs by PCR amplification with primer pairs flanking the targeted sites. Junction fragment amplicons of the expected size (bracket) were detected in eight of eight 4 dpf embryos that had been injected simultaneously with alb_2 and alb_3 dgRNPs and in five of eight 4 dpf embryos injected with alb_2 and alb_4 dgRNPs. Junction fragments recovered from starred embryos were sequenced and reported in Figure S4. See also Figures S4–S7. |
Reprinted from Developmental Cell, 51(5), Hoshijima, K., Jurynec, M.J., Klatt Shaw, D., Jacobi, A.M., Behlke, M.A., Grunwald, D.J., Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish, 645-657.e4, Copyright (2019) with permission from Elsevier. Full text @ Dev. Cell