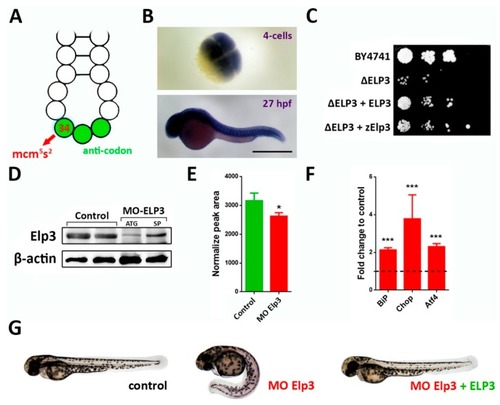
Elp3 morphants present a ventrally-curved tail. (A) Schematic representation of the anti-codon loop of a tRNA, indicating position 34 (red arrow) where 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) is located. Anti-codon is depicted in green. (B) In situ hybridization of Elp3 mRNA in 4 cell (upper) and 27 hpf (lower) embryos. Scale bar 0.7 mm. (C) Saccharomyces cerevisiae mutants for ELP3 (ΔELP3) were transformed with an expression plasmid that contained zebrafish Elp3 (zElp3). BY4741 was used as control. Image representative of three independent experiments. (D) Western blot detecting Elp3 in 48 hpf samples from control and morphant embryos (MO-Elp3); β-actin was used as loading control. Samples from embryos injected with translation- (ATG) and splicing-blocking (SP). (E) Liquid chromatography-mass spectrometry (LC-MS) analysis of tRNA modification. Peak area values for mcm5s2U are shown normalized to the of purified tRNA for each sample (n = 3, * p < 0.05). (F) qPCR for UPR targets BiP, Chop and Atf4. Fold change in mRNA levels comparing morphant embryos to controls; t-test was performed for statistical analysis. *** p < 0.005). (G) Representative images of control, morphants and MO + ELP3 mRNA injected embryos by 48 hpf (images representative of at least six independent experiments, in which at least 100 embryos were analyzed).
|

