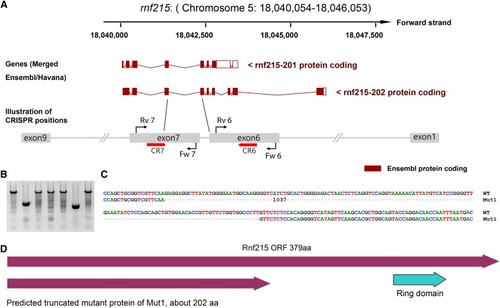
LKO mutations induced by multiple gRNAs targeting the rnf215 gene in zebrafish. Illustration of gRNAs that are against rnf215 exon 6 and exon 7 based on Ensemble annotation. Red bars with arrows indicate the positions of gRNAs against the rnf215 coding DNA. Red arrows match the gRNA directions. The lengths of illustrated exons are not proportional to the real size of corresponding exons. Black lines and red arrows represent primers and their directions. (B) Representatives of positive adult F1 fish by PCR with primers Fw 6 and Rv 7 that bind to exon 6 and exon 7, respectively. The top band was amplified from wildtype and the lower small band was amplified from the LKO mutant. Among the 48 fish, 3 fish were identified to have large deletions between exon 6 and exon 7 (also see Table 1). Each lane is corresponding to an F1 adult fish. (C) The exon 6 – exon 7 deletion (1,037bp) was confirmed by Sanger sequencing. 1,037bp is the size of the omitted region including the dashes. (D) Predicted zebrafish Rnf215 wildtype and mutant proteins based on the mutation positions. Protein domains were based on cBioPortal and SMART annotations. Only wildtype protein from the longest transcript is shown here. Mutant protein sizes were calculated by ORF finder.
|

