Fig. 7
- ID
- ZDB-FIG-200311-9
- Publication
- Farkas et al., 2019 - Discovering the chloride pathway in the CFTR channel
- Other Figures
- All Figure Page
- Back to All Figure Page
|
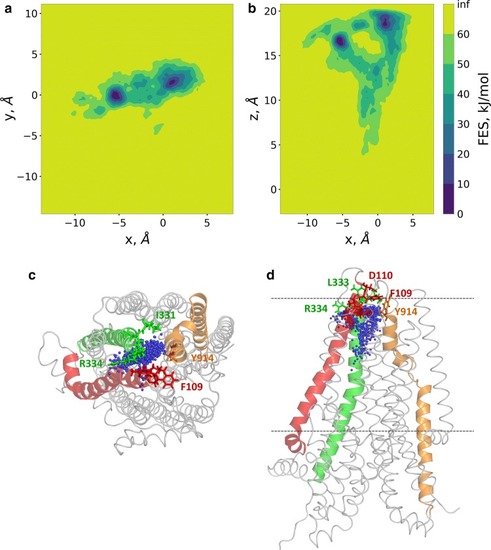
Two routes towards the extracellular regions were detected by metadyanamics simulations. Well-tempered metadynamics simulations were performed to identify potential exit routes of chloride ions and the amino acids narrowing this region of the pathway. The distance between the center of mass of four Cα atoms (a.a. 95, 347, 924, and 1141) and Cl− #2 was used as a reaction coordinate (collective variable, CV). 2D free energy surfaces were calculated along the |