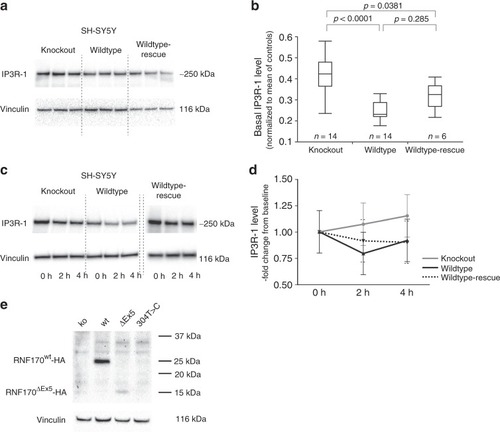
Effect of RNF170 mutations on IP3R-1 degradation and abundance in neuronal cells. a, b Immunoblot analysis of IP3R-1 in wildtype and knockout SH-SY5Y cells (SH-SY5Y(RNF170wt)/n = 14 biologically independent samples; SH-SY5Y(RNF170ko)/n = 14 biologically independent samples) and after re-expression of RNF170 in a knockout background (SH-SY5Y(RNF170ko(wt-HA))/n = 6 biologically independent samples). SH-SY5Y(RNF170ko) cells demonstrate significant accumulation of IP3R-1 (ko: mean 0.437 ± 0.133; wt: mean 0.247 ± 0.043) that can be rescued by re-expression of RNF170 (rescue: mean 0.318 ± 0.066; Tukey–Kramer HSD, two-sided). In the quantile blot, boxes indicate the 1st and 3rd quartile and median (center line); whiskers depict the 1st/3rd quartile ± 1.5* interquartile range. c, d IP3R-1 was activated by carbachol stimulation in neuronal SH-SY5Y cells, including wt and CRISPR/Cas9 generated RNF170 knockout cell lines (SH-SY5Y(RNF170wt), SH-SY5Y(RNF170ko) as well as SH-SY5Y cells stably expressing wildtype HA-tagged RNF170 in a knockout background (SH-SY5Y(RNF170ko(wt-HA)). IP3R-1 levels were quantified by western blot at baseline (t = 0 h) and 2 h/4 h after stimulation. Neither the effect of the genotype on IP3R-1 degradation (wt vs. ko: p = 0.1806; repeated measures full-factorial analysis) nor the interaction between genotype and time was significant (wt vs. ko, genotype*time: p = 0.1956; repeated measures full-factorial analysis). Nine independent biological replicates were examined per genotype. Means and standard deviations are shown for each data point. e Expression of episomally expressed HA-tagged RNF170 was analyzed by immunoblot in SH-SY5Y cells
|

