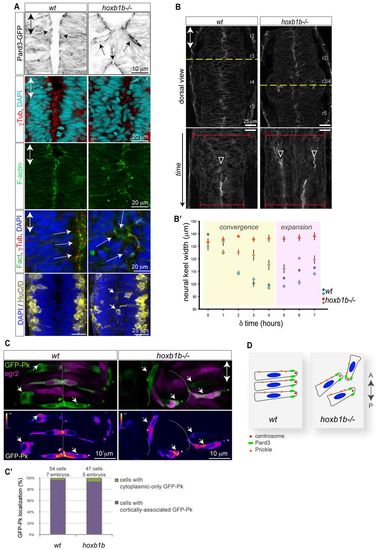
Normal cell polarity but disrupted convergence in the hoxb1b-/- neural keel. (A) Normal neuroepithelial polarity at the single-cell level in hoxb1b-/- based on sub-apical Pard3-GFP (black) at 21 hpf, apical centrosomes (γ-tubulin, red) and apically enriched F-actin (green), all analyzed at 19 hpf. Neuronal differentiation occurs at wild-type levels but generates HuC/D-positive neurons (yellow) in abnormal medial postitions in hoxb1b-/- at 20 hpf. (B) Dynamics of neural keel convergence in time-lapses of wild-type and hoxb1b-/- transgenic Gt(Ctnna-Citrine)ct3a embryos with multiphoton imaging. Upper panels show a single z-section in dorsal view one-third of the way through the time lapse, corresponding to about the 10-somite stage. Lower panels: kymograph of the position marked by a yellow dotted line, beginning at the 3-somite stage (11 hpf) and continuing until the 18 somite stage (18 hpf). A single lumen appears at the midline in wild type (arrowhead) whereas a duplicated midline appears at about the same stage in hoxb1b-/-. The width of the neural keel is depicted as red brackets. (B2) Quantification of neural keel width measured at three positions in r3/4 over the period in the kymograph. Convergence is significantly slowed in hoxb1b-/- compared with wild-type siblings (n=2 embryos for each genotype). Data are mean ± s.e.m. (C) Assessment of planar cell polarity by GFP-Prickle (green) localization at 18 hpf. Tg(egr2b:KalTA4) transgene (purple) is included to identify rhombomere 3. Mosaically expressed GFP-Pk localizes in puncta, which in wild type lie on anterior progenitor cell membranes (white arrows). In hoxb1b-/-, GFP-Pk localizes to puncta that lie on one side of mis-oriented progenitor cells. Lower panels show GFP-Pk signal presented in pseudocolors. (C2) Quantitative analysis of GFP-Prickle subcellular localization. (D) Schematic of cell polarity in hoxb1b-/-. The anterior-posterior animal axes are marked by double arrows in all panels.
|

