Fig. 2
- ID
- ZDB-FIG-080417-40
- Publication
- Davidson et al., 2003 - Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon
- Other Figures
- All Figure Page
- Back to All Figure Page
|
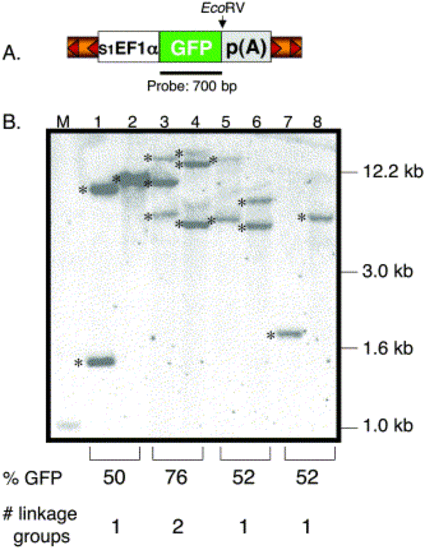
Southern blot analysis of transgenic zebrafish. (A) Structure of the pT2 transposon depicting the relative position of the GFP probe and EcoRV site. EcoRV cuts once in the pT2 transposon and NsiI is absent in the vector. (B). The EcoRV- or NsiI-digested genomic DNA was hybridized with the GFP-specific probe. Lanes 1, 3, 5, and 7 were digested with EcoRV and lanes 2, 4, 6, and 8 with NsiI. M, DNA marker lane. Shown are 6 independent insertions (*) from 3 individual F1 lines (lanes 3 and 4, lanes 5 and 6, lanes 7 and 8). The predicted number of linkage groups is based on the GFP expression data from the F1 outcrosses. One transgenic line previously shown to be carrying a single T-insertion was used as a control (lanes 1 and 2). |
Reprinted from Developmental Biology, 263(2), Davidson, A.E., Balciunas, D., Mohn, D., Shaffer, J., Hermanson, S., Sivasubbu, S., Cliff, M.P., Hackett, P.B., and Ekker, S.C., Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon, 191-202, Copyright (2003) with permission from Elsevier. Full text @ Dev. Biol.