Fig. 1
- ID
- ZDB-FIG-070919-12
- Publication
- Mann et al., 2006 - Comparison of neurolin (ALCAM) and neurolin-like cell adhesion molecule (NLCAM) expression in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
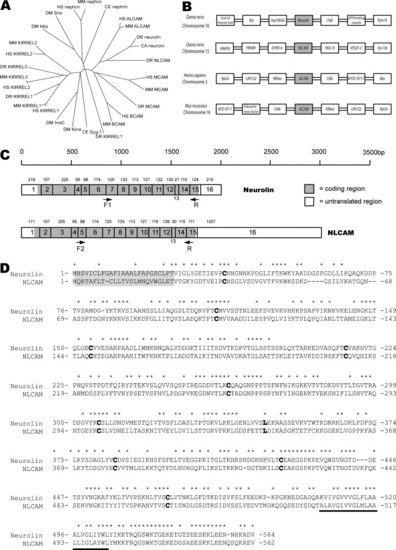
Family relationship, sequence and structural comparison of neurolin and NLCAM. (A) Phylogenetic tree of the ALCAM-group of proteins. The following families of proteins are identifiable: the ALCAM family, with the ALCAM- (including neurolin and NLCAM), BCAM- and MCAM-sub-families; the KIRREL-family; and the Nephrin-family. Duf/IrreC appear most closely related to KIRREL proteins. For species and gene abbreviations see Section 2. (B) The syntenic organisation of neurolin, NLCAM and ALCAM genes in zebrafish, human and mice, demonstrates partial duplication of the neurolin locus in vertebrates. (C) The predicted gene structure of neurolin and NLCAM mRNA is highly conserved suggesting a gene duplication. mRNAs shown approximately to scale. Exon size in base pairs (bp) is shown above each exon, and has been experimentally determined for all coding regions. The approximate location of probe PCR primers is shown. (D) The aligned protein sequences with identical residues marked with asterisk (*) above. The N-terminal signalling sequence (grey box) and transmembrane region (underlined) flank the cysteine (and solitary leucine) residues that define the Ig-like domains (bold). |
Reprinted from Gene expression patterns : GEP, 6(8), Mann, C.J., Hinits, Y., and Hughes, S.M., Comparison of neurolin (ALCAM) and neurolin-like cell adhesion molecule (NLCAM) expression in zebrafish, 952-963, Copyright (2006) with permission from Elsevier. Full text @ Gene Expr. Patterns