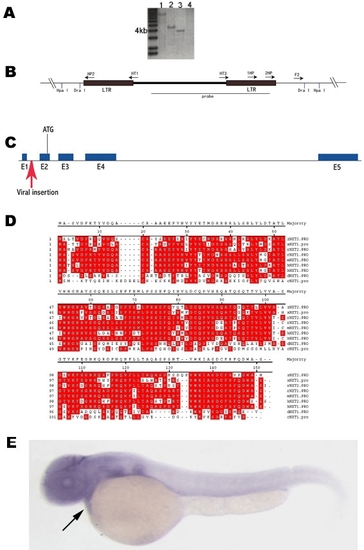
Identification of NXT2 as a candidate gene responsible for the heart defect. Southern blot analysis shows that a 4.3 kb (digested by Hpa I; A, lane 2) and a 3.8 kb (digested by Dra I; A, lane 3) are detectable using a specific viral DNA probe (B). Lanes 1 and 4 (A) are positive and negative control, respectively. DNA fragments corresponding to 3.8 kb / Dra I and 4.3 kb / Hpa I restriction were gel-purified and used for inverse PCR to isolate junction genomic sequences with primers described in (B) and Methods. This resulted in identification of NXT2 gene, which consists of five exons (C) and encodes 143 putative amino acids (D). The provirus is inserted within the first intron (red arrow) upstream of the putative initiation codon ATG in exon 2 (C). Alignments of protein sequences from various organisms revealed that zebrafish NXT2 shares 73.9%, 72.5%, 71.6%, 70.2%, 70.9%, 68.8%, 43.6% and 34.6% amino acid homology with that of rat NXT2, human NXT2, human NXT1, rat NXT1, mouse NXT1, Xenopus NXT1, Drosophila NXT1 and C. elegans NXT1, respectively (D). Identical residues are shown in red boxes. Zebrafish NXT2 is ubiquitously expressed at 2 dpf as shown by RNA whole mount in situ hybridization (E, arrow points to heart). E: lateral view, anterior to the left. zNXT2: zebrafish NXT2; hNXT1: human NXT1; hNXT2: human NXT2; mNXT1: mouse NXT1; rNXT1: rat NXT1; rNXT2: rat NXT2; cNXT1: C. elegans NXT1; dNXT1: Drosophila NXT1 and xNXT1: Xenopus NXT1.
|

