- Title
-
Transposon insertion causes ctnnb2 transcript instability that results in the maternal effect zebrafish ichabod (ich) mutation
- Authors
- Varga, Z., Kagan, F., Maegawa, S., Nagy, Á., Okendo, J., Burgess, S.M., Weinberg, E.S., Varga, M.
- Source
- Full text @ BBA Gene Regulatory Mechanisms
|
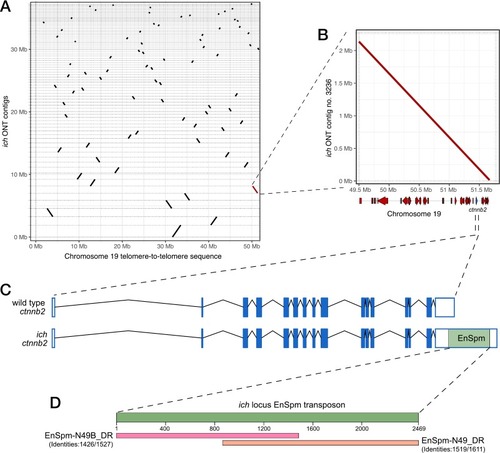
Long-read sequencing identifies an EnSpm transposon in the 3’UTR of ctnnb2 in ich animals. (A) Dotplot representation of different ich contigs that could be aligned to the “telomere-to-telomere” sequence of the zebrafish chromosome 19. The highlighted (red) contig contains the genomic region of the ctnnb2 gene. (B) Dotplot comparison of the relevant ich contig and the telomeric portion of chromosome 19 reveals no genomic rearrangement in this region in ich animals. Gene position schematics are visible along the horizontal axis. (C) Comparison of wild-type and ich-specific ctnnb2 sequences showing the position of the EnSpm-type transposon insertion. (D) BLAST results of the ich-specific transposon in the FishTEDB [40]. |
|
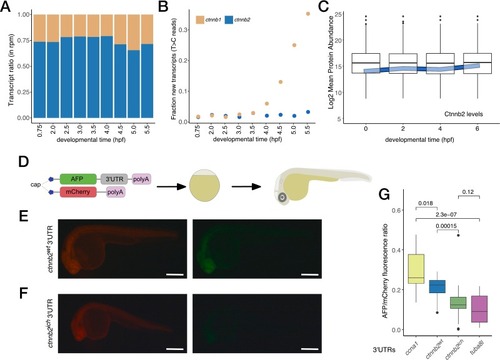
High stability of maternal ctnnb2 transcripts is disrupted by 3’UTR transposon insertion. (A) Transcriptome analysis of maternal ctnnb1 and ctnnb2 mRNAs shows a heightened abundance of maternal ctnnb2 transcripts throughout early development (rpm – reads per million bp, ctnnb1 transcripts are shown in beige, ctnnb2 transcripts in blue). (B) Metabolic labelling suggests active transcription of ctnnb1 after ZGA, but no expression of ctnnb2. (Data for panels A and B are derived from [41]. (C) Maternal Ctnnb2 protein levels are also very stable during early stages of development as compared to mean protein abundances at these timepoints. (Data from [43]. (D) Experimental scheme used to test the effect of 3’UTRs on the stability of mRNAs. In vitro transcribed AFP mRNAs with different 3’UTRs are combined with mCherry transcripts with invariant 3’UTRs and injected into 1-cell stage zebrafish embryos. Fluorescence intensity for both AFP and mCherry is measured at 24 hpf. (E, F) Typical fluorescence observed in embryos injected with AFP carrying ctnnb2wt or ctnnb2ich 3’UTRs, respectively. (Scale bars: 0.5 mm) (G) Relative fluorescence intensities observed in embryos injected with different 3’UTRs compared to stable and unstable controls (ccna1 and tuba8l 3’UTRs, respectively). (Pairwise p values were calculated with the Kruskal-Wallis test.) |
|
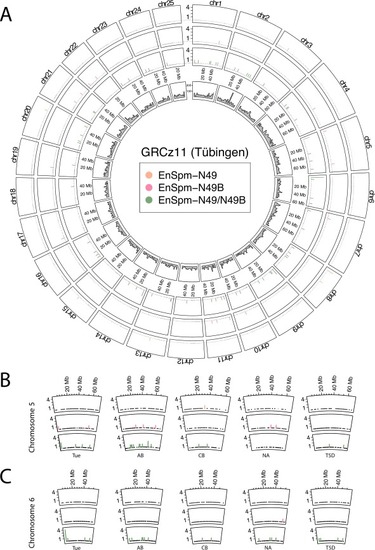
The position of EnSpm transposons observed in this study vary across zebrafish genomes. (A) Genomic positions of EnSpm-N49 (orange), EnSpm-N49B (pink) and EnSpm-N49/N49B (green) transposons in nonoverlapping 2-Mbp windows across the nuclear chromosomes of the Tübingen (Tue) GRCz11 reference genome. Inner circle shows the coverage of protein-coding genes on the same chromosomes. (B, C) Genomic positions of the observed transposons show variability across the genomes of multiple wild-type isolates for chromosomes 5 (B) and 6 (C). (Y axis denotes copy numbers in the 2-Mbp windows. Strain abbreviations can be found in Table 3. For other chromosomes see Supplementary Fig. 4.) |
Reprinted from Biochimica et biophysica acta. Gene regulatory mechanisms, , Varga, Z., Kagan, F., Maegawa, S., Nagy, Á., Okendo, J., Burgess, S.M., Weinberg, E.S., Varga, M., Transposon insertion causes ctnnb2 transcript instability that results in the maternal effect zebrafish ichabod (ich) mutation, 195104195104, Copyright (2025) with permission from Elsevier. Full text @ BBA Gene Regulatory Mechanisms