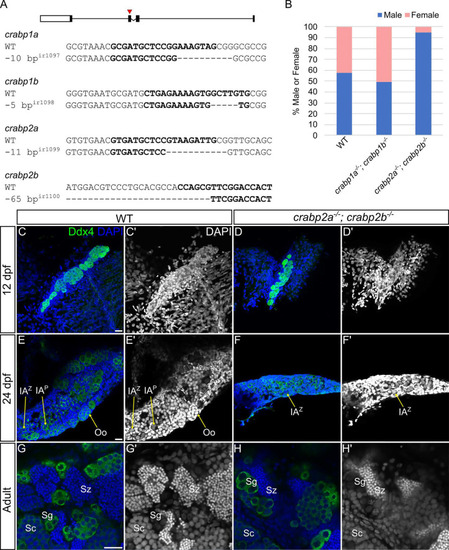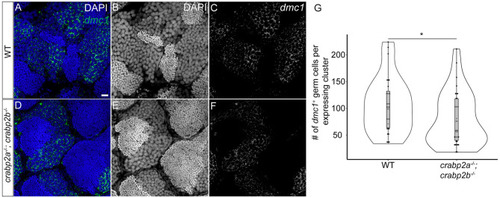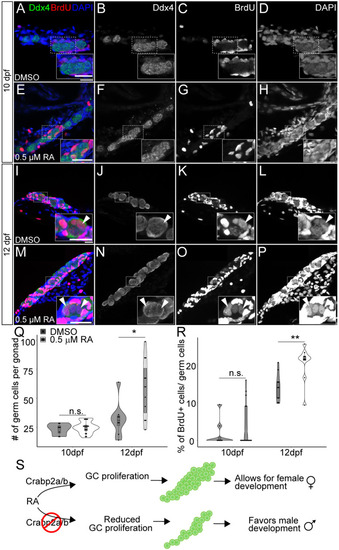
Retinoic acid promotes germ cell proliferation during gonad development. (A-H) Representative confocal z-projections of BrdU incorporation between 9 and 10 (A-H), and 11 and 12 (I-P) dpf in gonads of DMSO vehicle- (A-D,I-L) or 0.5 µM RA- (E-H,M-P) treated animals. Anti-Ddx4 labeled germ cells (GCs) are green; BrdU-labeled proliferating cell nuclei are red (A,E,I,M). (B,F,J,N) Grayscale Ddx4; (C,G,K,O) BrdU; (D,H,L,P) DAPI. Insets show magnified views of regions of interest in GCs (outlined). Arrowheads indicate GC nuclei showing BrdU incorporation. (Q) Violin, and box and whisker plots depicting the GC number per gonad at 10 and 12 dpf. Each datapoint represents the total GC number in one gonad. At 10 dpf, DMSO- and 0.5 µM RA-treated animals were similar (P=0.4930). At 12 dpf, DMSO-treated animals had significantly fewer GCs than 0.5 µM RA-treated animals (P=0.0182). (R) Violin, and box and whisker plots depicting percentages of proliferating GCs at 10 and 12 dpf. Each datapoint represents BrdU+ GC numbers per gonad. At 10 dpf, most DMSO- and 0.5 µM RA-treated animals showed no BrdU+ GCs (P=0.4209). At 12 dpf, DMSO-treated animals had significantly fewer BrdU+ GCs compared to 0.5 µM RA-treated animals (P=0.0044). An unpaired two-tailed t-test was used to test for significance (*P<0.05; **P<0.01; n.s., no significance). (S) Schematic for proposed roles of RA and Crabp2 proteins in regulating GC development and sex differentiation. Violin plots: DMSO treatments, gray bars; 0.5 µM RA, white bars. The box represents the interquartile range (IQR) where 50% of the data points are present. The height of the box is inversely proportional to the clustering of the measurements. Outliers are present outside the box and quartiles. The horizontal line in the box plot represents the median. Scale bars: 20 μm. DMSO: 10 dpf, n=3 larvae; 12 dpf, n=7 fish. RA treated: 10 dpf, n=3 larvae; 12 dpf, n=8 fish.
|