|
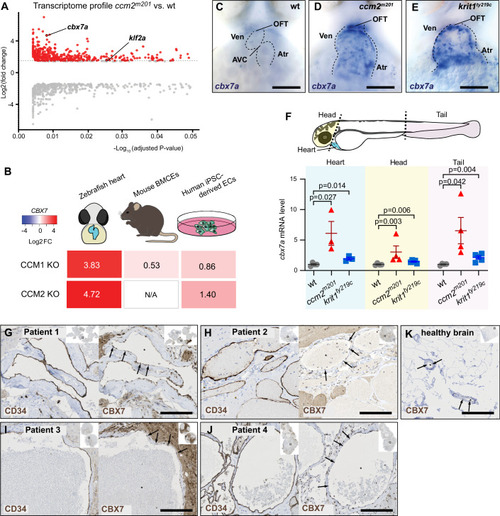
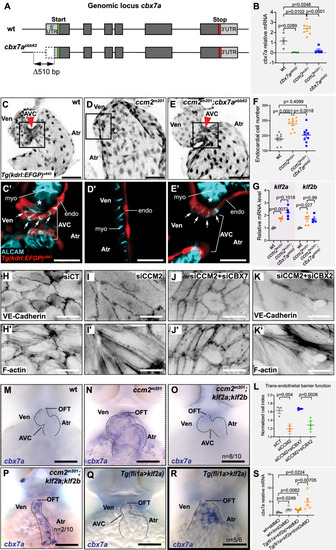
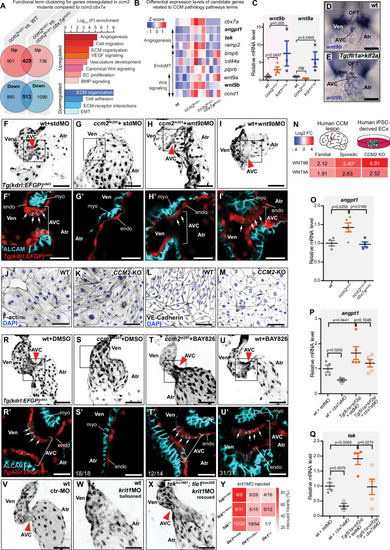
Cbx7a/CBX7 suppresses CCM-associated phenotypes in zebrafish ccm2m201 mutants and is controlled by Klf2a and blood flow. (A) Schematic representation of the cbx7a genomic locus in zebrafish. The cbx7apbb62 allele comprises a 510 bp deletion 80 bp upstream of the starting ATG (green bar). Areas in dark gray indicate the coding region and light gray areas highlight 5′ and 3′ untranslated regions (UTR). (B) Quantification of cbx7a mRNA levels show that its expression is completely depleted in cbx7apbb62 mutants (p = 0.0289) and in ccm2m201;cbx7apbb62 double mutants (p = 0.0248). In ccm2m201 mutants, cbx7a mRNA levels are upregulated (p = 0.0102). Four to six replicates per group, pooled 8–10 embryos per each replicate. Statistical testing is based on one-way ANOVA with Tukey’s multiple comparisons test (s.e.m. bars indicated). (C–E) Endocardium at 56 hpf, marked by Tg(kdrl:EGFP)s843 expression in wild-type (wt) (C), ccm2m201 (D), and ccm2m201;cbx7apbb62 double mutants (E). The endocardium forms the AVC (red arrowhead) between the ventricle and the atrium at 56 hpf (C). Endocardial cells of the AVC are marked by the expression of the cell junctional protein ALCAM (C’). The asterisk indicates the location of cardiac jelly between the myocardium and endocardium (C’). In ccm2m201 mutants, there is no AVC constriction (D) and endocardial cells lack ALCAM expression, and the cardiac jelly is markedly reduced (D’). The endocardial defects in zebrafish ccm2m201 mutants were suppressed when Cbx7a is genetically depleted (E). ccm2m201;cbx7apbb62 double mutants form an AVC constriction (E), and ALCAM expression (E’) and cardiac jelly (E’, asterisk) are restored. (F) Quantifications demonstrate that ccm2m201 mutants have higher endocardial cell numbers (p < 0.0001) and there is a normalization in ccm2m201;cbx7apbb62 double mutants (p = 0.4099 compared to wild-type; p = 0.0018 compared to ccm2m201). Each data point represents endocardial cell numbers counted from a single heart. Statistical testing is based on the student’s T-test (s.e.m. bars indicated). (G) Expression levels of klf2a and klf2b mRNA are elevated in ccm2m201 and ccm2m201;cbx7apbb62 double mutants. Shown are quantifications of whole embryos by qRT-PCR. mRNA levels of klf2a and klf2b are increased in ccm2m201 mutants as compared to wild-type (p = 0.0072 and p = 0.027, respectively) and do not normalize in ccm2m201;cbx7apbb62 double mutants (p = 0.1018 and p = 0.99 for klf2a and klf2b, respectively; n = 4–5 replicates per each group; 8–10 embryos were collected for each replicate). Statistical testing is based on student’s T-test between mutants and corresponding wild-type siblings (s.e.m. bars indicated). (H–K) Representative images of monolayers of HUVECs silenced for control (siCT) (H, H’), CCM2 (I, I’), co-silencing of CCM2 and CBX7 (J, J’), and co-silencing of CCM2 and CBX2 (K, K’). Phenotypes of CCM2-depleted HUVECs show an elongated morphology with VE-cadherin at cell-cell junctions and increased stress fiber formation (I, I’) as compared to controls (H, H’). Restoration of cell morphology with cortical actin and reduced stress fibers is observed upon knock-down of CBX7 (J, J’), but not CBX2 (K, K’). (L) Quantification of the normalized impedance of HUVEC monolayers 24 h after serum starvation. Knockdown of CCM2 in HUVECs causes a reduction of barrier function, which is restored upon co-depletion of CBX7, but not with co-depletion of CBX2. There are three replicates per each group and statistical testing is based on one-way ANOVA with Tukey’s multiple comparisons test (s.e.m bars indicated). (M–R) Representative images of cbx7a expression detected by whole-mount in situ hybridization in the zebrafish heart at 56 hpf. (M) cbx7a expression is low in wild-type and (N) expanded throughout the entire endocardium in ccm2m201 mutants. (O) Higher levels of cbx7a mRNA in ccm2m201 mutants are reduced in ccm2m201; klf2ash317; klf2bpbb42 triple mutants with normal heart morphology (n = 8/10 triple mutants with reduced cbx7a mRNA expression) and elevated in triple mutants in which the cardiac morphology has not been restored (P). In Tg(fli1a:Gal4FF)ubs3;Tg(UAS:klf2a)ig1 double transgenic embryos [Tg(fli1a>klf2a)], in which blood flow is present, mRNA levels of cbx7a remain low (Q). Upon endothelial-specific overexpression of Klf2a and concomitant loss of blood flow, cbx7a mRNA levels are elevated (R). (S) Quantification of cbx7a mRNA levels by qRT-PCR in Tg(fli1a>klf2a) embryos that also lack blood flow (tnnt2a MO against sarcomeric protein Tnnt2a) or with normal cardiac function (standard morpholino, std MO). There are five replicates per each group and each replicate is based on 8–10 pooled embryos. Statistical testing is based on student’s T-test (s.e.m. bars indicated). AVC atrioventricular canal, endo endocardium, myo myocardium, OFT outflow tract, Ven ventricle, Atr atrium. Red arrowheads indicate the location of the AVC and white arrows indicate the presence of ALCAM expression in AVC endocardial cells. Experiments in zebrafish were done in three biological replicates. Scale bars are (M–R) 100 µm, (C–E, H–K, H’–K’) 50 µm, (C’–E’) 20 µm. Source data are available online for this figure.
|