- Title
-
An atypical form of 60S ribosomal subunit in Diamond-Blackfan anemia linked to RPL17 variants
- Authors
- Fellmann, F., Saunders, C., O'Donohue, M.F., Reid, D.W., McFadden, K.A., Montel-Lehry, N., Yu, C., Fang, M., Zhang, J., Royer-Bertrand, B., Farinelli, P., Karboul, N., Willer, J.R., Fievet, L., Bhuiyan, Z.A., Kleinhenz, A.L., Jadeau, J., Fulbright, J., Rivolta, C., Renella, R., Katsanis, N., Beckmann, J.S., Nicchitta, C.V., Da Costa, L., Davis, E.E., Gleizes, P.E.
- Source
- Full text @ JCI Insight
|
Pathogenic variants in RPL17 cause Diamond-Blackfan anemia. (A) 4-generation Swiss pedigree (family 1) harboring a heterozygous c.217-3C>G variant in RPL17. Filled shapes, affected individuals; unfilled shapes, unaffected individuals; orange outline of shapes, exome sequencing was performed on DNA from that individual; “+”, WT allele; black triangle, index case. Individuals for whom no genotype is indicated means that a DNA sample was unavailable. (B) RPL17 mutation carriers of family 1 display digit phenotypes including fifth finger hypoplasia (1-I-1, 1-II-2, 1-III-3, 1-II-1, 1-II-4, and 1-III-5) and absent thumbs (1-III-6 and 1-IV-2). (C) Family 2 harbors a heterozygous c.452delC RPL17 variant and displays incomplete penetrance. Colors and symbols are the same as indicated in panel A. (D) Schematic of the WT human RPL17 locus (bottom) and mRNA (top). Black boxes, coding exons; white boxes, untranslated regions; solid black lines, introns; green “AUG”, start codon; red “UAA”, stop codon; and variants identified in this study, red (pathogenic) or green (benign) asterisks. (E) RPL17 variants identified in each of families 1 and 2 produce aberrant mRNA transcripts; blue bars in the schematic highlight the site of alteration. (F) RT-PCR products separated on agarose gel indicating that c.217C>G results in an in-frame deletion of exon 4. (G) Immunoblot of RPL17 protein (21.4 kDa) in LCLs derived from cases (gray) and unaffected individuals (black bold) from families 1 and 2. |
|
Characterization of erythroid maturation defects in family 1 from cells cultured in vitro. (A) Cell growth curves during erythroid cell differentiation; 1-II-2, 1-II-4, 1-III-3, and 1-III-5 harbor the RPL17 c.217-3C>G variant; 1-II-5 is a healthy control (married-in spouse); D, day of culture. (B) Time course of CD34 and CD36 labelling. FACS analysis at D7 showed no consistent change in the percentage of BFU-e (CD34+/ CD36–) or CFU-e (CD34–/ CD36+) progenitor cells with RPL17 variants. The gradual loss of CD36 labelling from D12 to D15, indicative of terminal differentiation stages, occurred with similar kinetics in cells from both affected and unaffected individuals. Profound cell death of sample 1-III-3 prohibited study beyond D9. (C) Quantification of GPA+ cells (erythroid specific marker) by FACS from D7 to D15. (D) Erythropoietic differentiation was assessed by co-detection of α4-integrin and Band3 in GPA+ cells. Increase in Band3 labelling, a marker of terminal erythroid cells, is paralleled by a decrease of α4-integrin during erythroid differentiation (35). (E) Quantification of the early apoptotic marker, Annexin V from D7 to D15 shows no significant apoptosis in patient-derived cells compared with control cells. |
|
Zebrafish models of rpl17 ablation display anemia and craniofacial patterning defects. (A) Representative lateral views of gata1:dsRed larvae imaged at 3 days postfertilization (dpf). Fluorescent signal, indicative of erythroid precursors and erythrocytes, was quantified in a consistently sized region of interest (red box) located posterior of the cloaca on the ventral side of controls, F0 mutants, and MO-injected larvae. Anterior, left; posterior, right. (B) Quantification of dsRed+ cells (erythroid cells) in the region of interest (see panel A) in F0 mutant or morphant larvae at 3 dpf. n = 20–40 larvae/batch, repeated at least twice. (C) Representative ventral views of –1.4col1a1:egfp larvae imaged at 4 dpf. Fluorescent signal was assessed for cartilage patterning defects by measuring the angle of the ceratohyal (ch) cartilage (dashed lines). Ceratobranchial (cb) arches were also dysplastic and reduced in number compared with controls. Anterior, left; posterior, right. (D) Quantification of the ch angle in F0 mutant and morphant larvae at 4 dpf; n = 16–32 larvae/batch, repeated at least twice. mRNA encodes predicted proteins p.A73_K105del and p.(T151Rfs*25) corresponding to Family 1 and Family 2, respectively. mRNA coding for p.Q56L is present in public databases (rs753489644; gnomAD browser), and scores as a benign variant in this assay. In panels B and D, ends of the whiskers are set at 1.5 times the interquartile range (IQR) above the third quartile and below the first quartile, respectively. Black dots, minimum and maximum outliers; *P < 0.05; **P < 0.01; **** P < 0.0001; (Kruskal-Wallis with Dunn’s multiple comparisons test). |
|
Analysis of ribosome synthesis show defects in rRNA maturation. (A) Total RNAs extracted from LCLs of DBA cases (gray) or unaffected individuals (black) were analyzed by Northern blot with probes ITS2, 5′-ITS1, 18S, and 28S (positions of the probes in panel C). The ratios of 28S to 18S rRNAs were quantified, normalized to the mean value obtained for controls, and are displayed as log values. (B) Detection of the cryptic pre-rRNA species 36S, 36S-C, and 32.5S with the ITS1-5.8S probe. The intensity profiles shown for the ITS1-5.8S probe were normalized relative to the levels of 28S rRNA. (C) Schematic representation of the pre-rRNAs derived from the 47S primary ribosomal transcript in human cells by endonucleolytic cleavage (horizontal lines) and exonucleolytic processing (Pacman). Impairment of cleavage at site 2 leads to accumulation of the 36S and 36S-C cryptic precursors by direct ITS1 cleavage at site E. Domain C corresponds to a highly conserved domain in ITS1 that blocks exonuclease progression. The positions of the Northern blot probes are indicated below the 47S pre-rRNA and their sequences are listed in Supplemental Table 8. |
|
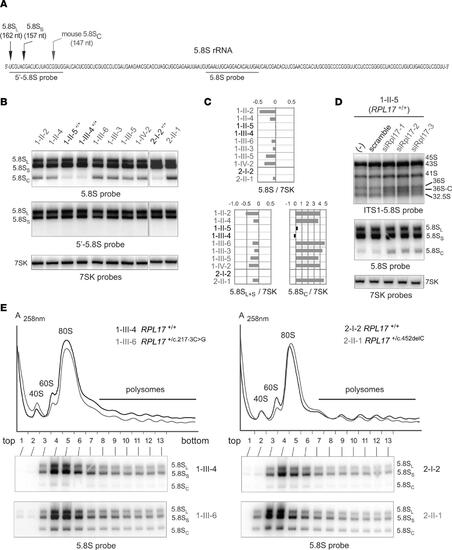
A very short form of 5.8S rRNA contributes to ribosome heterogeneity in RPL17+/mut cells. (A) Sequence of the human 5.8S rRNA with the 2 canonical 5′ ends and that of mouse 5.8SC rRNA previously determined (54), with the positions of the probes used in this figure. (B) 5.8S rRNA species separated on a 6% polyacrylamide gel (bottom) were identified with the 5.8S probe hybridizing to the core of the 5.8S rRNA, or with the 5′-5.8S probe complementary to the 5′-end of the 5.8S (A). The 7SK RNA was used as a loading control. (C) The ratios of the 5.8S rRNA species to 7SK snRNA (B) were quantified. Values were normalized to the mean value obtained for controls and are displayed as log2 values. (D) LCLs from unaffected individual 1-II-5 were treated with 3 siRNAs targeting RPL17 mRNA. Total RNA profiles analyzed on agarose (top) or polyacrylamide gels (bottom) were compared to those of untreated cells (–) or cells treated with a scramble siRNA. (E) Cytoplasmic fractions extracted from control (black) or case (gray) lymphoblastoid cell lines were analyzed by ultracentrifugation on 10%–50% sucrose gradients. Total RNAs from the gradient fractions were extracted and analyzed by Northern blot with the 5.8S probe. |
|
A specific translational response in LCLs of patients with DBA. (A) Experimental schematic. Cells from DBA cases or from healthy controls were lysed and treated with nuclease to degrade mRNA not protected by ribosomes. Ribosome-protected mRNA was then purified and deep sequenced to quantify translational activity. (B) Histogram showing the changes in ribosome density (ribosome profiling read density divided by RNA-Seq read density) in DBA cells relative to controls. The change in mRNA levels is also shown. All DBA cells carrying the RPL17 c.217-3C>G genotype are included. (C) Gene ontologies enriched in mRNAs that have suppressed or enhanced translation. The log2 change represents the mean change in total translation across all DBA cells, with P values calculated by bootstrapping. (D) Relationships in translational activity between WT and DBA cells. Similarities are calculated by Pearson’s correlation coefficient. |