- Title
-
CORRECTION: New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts
- Authors
- Maugars, G., Nourizadeh-Lillabadi, R., Weltzien, F.A.
- Source
- Full text @ Front Endocrinol (Lausanne)
|
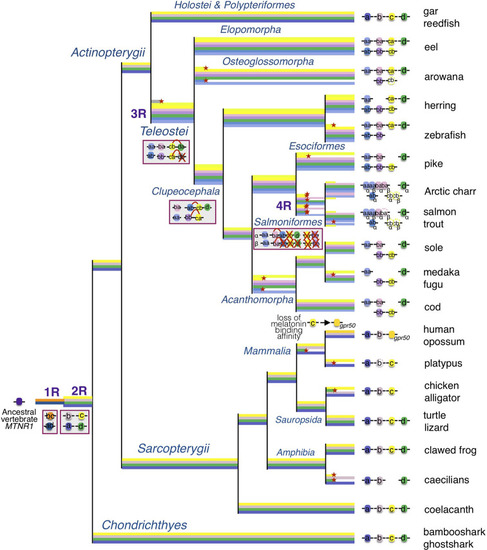
Evolutionary scenario of the melatonin receptors in vertebrates. This evolutionary scenario was developed on the basis of the phylogeny and synteny analyses presented in the original article. The four receptor genes are derived from duplication of an ancient |