- Title
-
CORRECTION: A miRNA catalogue and ncRNA annotation of the short-living fish Nothobranchius furzeri
- Authors
- Baumgart, M., Barth, E., Savino, A., Groth, M., Koch, P., Petzold, A., Arisi, I., Platzer, M., Marz, M., Cellerino, A.
- Source
- Full text @ BMC Genomics
|
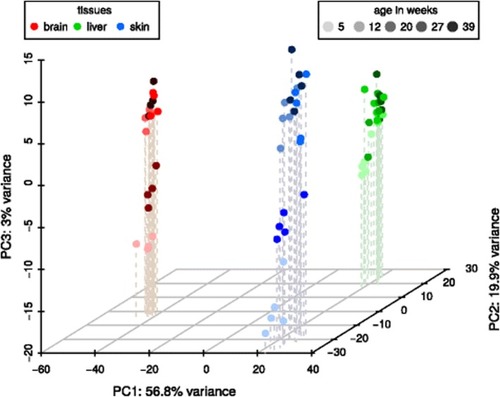
A three-dimensional PCA plot of the |
|
Annotation, expression profiles and prediction comparison for miR-499.We annotated the pre-miR-499 on sgr09, position 55,926,017– 55,926,134 and the two mature miRNAs at 55,926,048–55,926,069 and 55,926,085–55,926,106. The six methods used for miRNA detection are displayed, CID-miRNA was not able to detect this miRNA. Tools working independent of the small RNA-Seq data BLAST (cyan), Infernal (olive green) and goRAP (orange) vary in their annotation length. The latter two programs are based on covariance models, identifying mostly the complete pre-miRNA. The remaining two programs miRDeep* and Blockbuster are based on small RNA-Seq data (*) and therefore accurately annotate the mature miRNAs. MiR-499 is expressed weakly within |
|
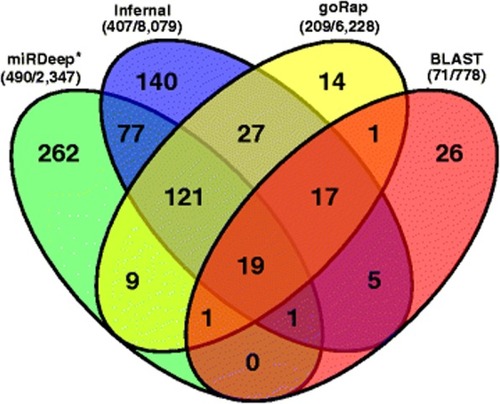
Venn diagram of predicted miRNA genes from four tools miRDeep*, Infernal, goRap and BLAST. Only 2 of the 33 candidates predicted by CID-miRNA overlapped with any of the other miRNA candidates. Nevertheless, all 33 candidates were selected as miRNAs after manual inspectations. The total number of miRNA predictions after and before applying any filtering step are shown in brackets for each tool |