- Title
-
FANCI functions as a repair/apoptosis switch in response to DNA crosslinks
- Authors
- Shah, R.B., Kernan, J.L., van Hoogstraten, A., Ando, K., Li, Y., Belcher, A.L., Mininger, I., Bussenault, A.M., Raman, R., Ramanagoudr-Bhojappa, R., Huang, T.T., D'Andrea, A.D., Chandrasekharappa, S.C., Aggarwal, A.K., Thompson, R., Sidi, S.
- Source
- Full text @ Dev. Cell
|
ICLs trigger PIDDosome signaling.
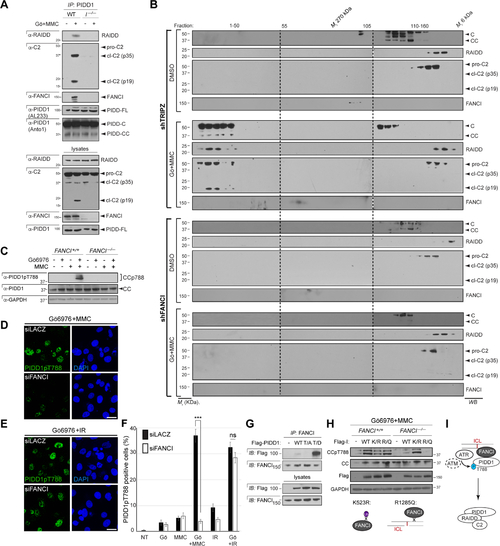
(A) HeLa cells stably expressing the indicated shRNAs, treated with or without Gö6976 and MMC (1 μM each) and harvested 24hrs post-MMC were analyzed by western blot. pro-C2, procaspase-2; p35, intermediate cleavage fragment; p19, mature product. FL, full-length; C, large C-terminal autocleavage product; CC, short, distal, C-terminal autocleavage product (see J). (B) HeLa cells of indicated genotypes stably expressing C2 Pro-BiFC and mCherry were treated with Gö6976 and MMC (5μM each) plus VD-OPH (20 μM) and fixed at 24 hours. Confocal images (5 μm sections) show C2 BiFC (yellow) and mCherry (red) expression. Scale bar, 20 μm. (C) Quantification of images as in (B) over 3 independent experiments, with data expressed as means +/− SD. **p < 0.01, two-tailed Student’s t-test. (D) HeLa cells stably expressing the indicated shRNAs were treated with or without Gö6976 and MMC (1 μM each). Cells were stained with the vital dye alamarBlue 72 hr post-treatment. Data are means +/− SD of 3 independent experiments. **p < 0.01, two-tailed Student’s t-test. (E) HeLa cells initially treated with ATM inhibitor (KU55933) and ATR inhibitor (ETP46464), 10 μM each, were treated with or without Gö6976 and MMC (1 μM each) or IR (10 Gy) after 1 hr. Cells were harvested 24hrs post-treatment and analyzed by western blot. (F-I) HCT116 cells of indicated FANCI genotypes and stably expressing shp53 were treated with Gö6976 and MMC (1 μM each) and harvested at 0 hrs, 9 hrs and 24 hrs post-MMC. Lysates (I) were immunoprecipitated with s-17 antibody to the PIDD1 N-terminus (F) or monoclonal anti-ATR (G) and anti-ATM (H) antibodies and analyzed by western blot. (J) Schematic diagram of full-length (FL) PIDD1 (910 a.a.) and autoproteolytic cleavage products PIDD1-N, PIDD1-C and PIDD1-CC. LRR, leucine-rich repeats; ZU-5, ZO-1 and Unc5-like domain; UPA, uncharacterized protein domain in UNC5, PIDD and Ankyrin, implicated in PIDD1 oligomerization (Janssens and Tinel, 2012); DD, death domain. (K) HeLa cells transfected with the indicated Flag-tagged PIDD deletion constructs were harvested 24hrs post-transfection. Flag IPs were analyzed by western blot. (L) Recombinant GST-PIDD1-CC proteins (WT and T788A phosphomutant, T/A) were incubated with Flag-ATR and His-ATRIP for an in vitro kinase assay. Reactions were analyzed by Coomassie staining and western blot. α-pSQ/TQ, polyclonal antibody to phosphorylated SQ or TQ motifs, such as T788Q789 in PIDD1 (Ando et al., 2012). *, non-specific band. (M) Schematic of PIDD1 phosphorylation by ATR and ATM in response to ICLs and DSBs. ATM plays a delayed role in the ICL response (dotted). |
|
FANCI interacts with PIDD1 and is essential for ICL-induced PIDDosome signaling.
(A) Venn diagram featuring FANCI as the sole ICL-repair protein identified in a mass spectrometry analysis of Flag-PIDD1 immunoprecipitates (Logette et al., 2011). (B) HeLa cells treated with or without Gö6976 and MMC (1 μM each) were harvested after 24 hrs and monoclonal PIDD1 IPs were analyzed by western blot. *non-specific band. (C) HeLa cells treated with or without Gö6976 and MMC (1 μM each) were harvested after 24hrs and monoclonal FANCI IPs were analyzed by western blot. (D) HeLa cells grown on coverslips and synchronized as in Table S1A were treated with or without Gö6976 and MMC (1 μM each), fixed 9 hrs post-MMC, stained with the indicated antibodies and visualized by confocal microscopy (0.8 μm sections are shown). Arrowheads mark colocalization areas. Scale bar, 20 μm. (E) Schematic diagram of FANCI (1328 a.a) highlighting the FANCD2 binding domain on its C-terminal tail (aa 1001–1328). ARM, Armadillo domain, α-α superhelix folds involved in protein-protein interactions. Leu, leucine-rich region. EDGE, EDGE motif. (F) Schematic diagram of PIDD1-FL (910 a.a.) (G) HeLa cells transfected with indicated Flag-FANCI constructs were harvested after 24 hrs and Flag IPs were analyzed by western blot. (H) HeLa cells transfected with the indicated Flag-PIDD1 constructs were harvested after 24 hrs and FANCI IPs were analyzed by western blot. (I) HCT116 cells of indicated FANCI genotypes and stably expressing shp53 were treated with Gö6976 and MMC (1 μM each) and harvested 24 hrs post-MMC. Lysates were analyzed by western blot. (J) Patient-derived FA-I 010191 fibroblasts reconstituted with FANCI or empty vector (e.v.) were treated and analyzed as in (I). (K) HeLa cells stably expressing the indicated shRNAs were treated with or without Gö6976 (1 μM), MMC (1 μM) or IR (10 Gy) and analyzed as in (I). (L) HeLa cells stably expressing the indicated shRNAs were treated with Gö6976 (0.1 μM) and MMC (0.05 μM). 14 days post-treatment, cells were fixed and stained with crystal violet. (M) Colonies from (M) of greater than 50 cells were counted. Data from 3 independent experiments expressed as means +/− SEM. *p < 0.05; **p < 0.01 ***p < 0.001, two-tailed Student’s t-test. (N) p53–/– zebrafish embryos of indicated fanci genotypes were treated with or without Gö6976 (1 μM) at 17 hrs post fertilization (hpf) and MMC (30 μM) 1 hr later and stained with the cell death marker acridine orange (AO) at 24 hpf. Boxed spinal-cord areas are magnified. Scale bar, 0.2 mm. As in human cells (L), zebrafish FANCI is required for death induction in embryos treated with MMC+Chk1i. (O) Quantification of spinal cord AO stains as shown in (N). Data were collected from 2 independent experiments with at least 5 embryos scored per condition in each and reported as means +/− SEM. **p < 0.01, two-tailed Student’s t-test. |
|
FANCI acts directly to enable ATR/ATM-mediated PIDD1 phosphorylation and PIDDosome formation.
(A) HCT116 cells of indicated FANCI genotypes and stably expressing shp53 were treated with Gö6976 and MMC (1 μM each), harvested 24 hrs post-MMC and monoclonal PIDD1 IPs were analyzed by western blot. I–/–, FANCI–/–. (B) HeLa cells stably expressing the indicated shRNAs were treated with or without Gö6976 and MMC (1 μM each), lysed 24 hrs after treatment and run on a S400 HiPrep 16/60 Sephacryl column (1 ml/min). An aliquot of each fraction was concentrated and analyzed by western blot. (C) HCT116 cells of indicated FANCI genotypes and stably expressing shp53 were treated with Gö6976 and MMC (1 μM each), harvested at 24 hrs post-MMC and analyzed by western blot. (P) (D) PC3 cells grown on coverslips and transfected with indicated siRNAs for 48hrs were treated with Gö6976 and MMC (1 μM each). Cells were fixed after 10hrs of treatment, stained using indicated antibodies and visualized by confocal microscopy (0.8 μm sections). Scale bar, 40 μm. (E) As in (D) but substituting IR (10 Gy) for MMC. Scale bar, 40 μm. (F) Quantification of images as in (D-E) over three independent experiments. Data shown as means +/− SEM. ***p<0.001 and ns, non-significant, two-tailed Student’s t test. (G) HeLa cells transfected with the indicated C-terminally Flag-tagged PIDD1 constructs were lysed at 24 hpt and FANCI IPs were analyzed by western blot. WT, wild-type; T/A, T788A; T/D, T788D. (H) HCT116 cells of indicated FANCI genotypes and stably expressing shp53 were transfected with indicated FANCI cDNAs and treated with Gö6976 and MMC (1 mM each) after 24 hrs. Cells were harvested 24hrs post-treatment and analyzed by western blot. Bottom, schematics of K523R (mono-Ub-deficient) and R1285Q (DNA binding-deficient). (I) Model for FANCI-mediated PIDDosome signaling (see text). |
|
ICL-induced PIDDosome signaling does not strictly require FA pathway function.
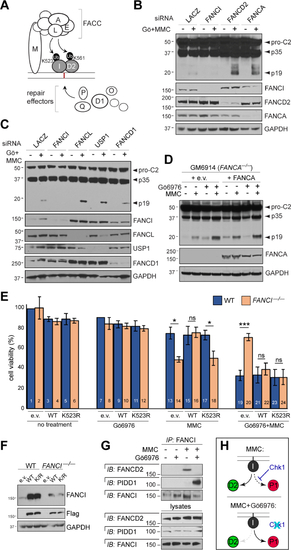
(A) Simplified diagram of the FA pathway. FACC, Fanconi core complex. (B-C) HeLa cells transfected with indicated siRNAs were treated with or without Gö6976 and MMC (1 μM each), harvested 24hrs post-treatment and analyzed by western blot. (D) Patient-derived GM6914 FANCA–/– fibroblasts reconstituted with empty vector or WT FANCA were treated with Gö6976 and MMC (1 μM each), harvested 24 hrs post-MMC and analyzed by western blot. (E) HCT116 cells of indicated FANCI genotypes and stably expressing shp53 were reconstituted with empty vector (e.v) or indicated Flag-FANCI constructs and treated with or without Gö6976 (1 μM) and MMC (0.1 μM). AlamarBlue was added 72 hrs post-treatment. Data are means +/− SD of 3 independent experiments. *p < 0.05; **p < 0.01 ***p < 0.001, two-tailed Student’s t-test. (F) Western blot from the experiment in (E) showing equal, near-physiologic expression of Flag-FANCI constructs. (G) HeLa cells synchronized as in Table S1A were treated with or without Gö6976 and MMC (1 μM each) and harvested after 9hrs. FANCI IPs were analyzed by western blot. (H) Schematic highlighting the effects of Chk1 (top) and Chk1i (bottom) on binding partner choice by FANCI in response to ICLs. |
|
Exclusive recruitments of FANCD2 and PIDD1 by FANCI enable a binary fate switch in response to ICLs.
(A) Schematic diagram of FANCI highlighting the overlapping FANCD2- and PIDD1- binding domains (D2BD and P1BD, respectively; see Figure 2G–H). (B) HeLa cells transfected with fixed or increasing amounts of expression vectors encoding HA-FANCI, Flag-PIDD1-FL and GFP-FANCD2, as indicated. Lysates were harvested 24 hrs post transfection and HA IPs were analyzed by western blot. (C) HeLa cells stably expressing the indicated shRNAs or transfected with indicated siRNAs were synchronized as in Table S1B, treated with or without Gö6976 and MMC (1 μM each) and harvested at 9 hrs post-treatment. FANCI IPs were analyzed by western blot (D) HeLa cells synchronized as in Table S1A were treated with or without Gö6976 and MMC (1μM each), harvested at indicated time points post-treatment, stained with phospho-Histone H3 antibody and analyzed by flow cytometry. (E) FANCI IPs from HeLa cells as in (D) were analyzed by western blot. hpt, hours post-treatment; int, interphase; M, mitosis. (F) HeLa cells transfected with indicated siRNAs and treated with or without MMC (1 μM) were harvested at 24 hrs. FANCI IPs were analyzed by western blot (G) HeLa cells synchronized as in Table S1A were treated with or without RO-3306 (10 μM) or nocodazole (200 ng/ml) 1 hr after release. Cells were then treated with Gö6976 (1 μM) and MMC (1 μM) and harvested 9 hrs later. FANCI IPs were analyzed by western blot. (H) Schematic summarizing the results in panels D-G. |
|
FANCI mounts a bidirectional fate switch that transduces failure of DNA repair into apoptosis, and vice versa.
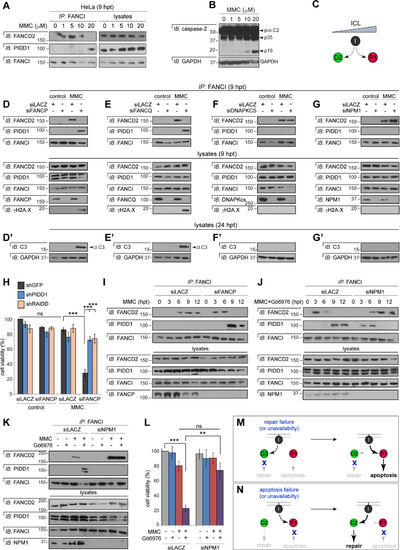
(A) HeLa cells treated with indicated doses of MMC were harvested at 9 hrs. FANCI IPs were analyzed by western blot. (B) HeLa cells treated with indicated doses of MMC were harvested at 24 hrs and analyzed by western blot. (C) Model derived from the results in panels A-B: FANCI switches from FANCD2 to PIDD1 with increasing ICL levels. (D-G) HeLa cells synchronized as in Table S1B were transfected with indicated siRNAs, treated with or without MMC (1 μM) and harvested 9 hrs post-MMC. FANCI IPs were analyzed by western blot. (D’ – G’) As above but harvested 24 hrs post-MMC and whole-cell lysates analyzed by western blot. cl.C3, cleaved caspase-3. (H) HeLa cells expressing the indicated shRNAs were transfected with indicated siRNAs, treated with or without MMC (0.1 μM) and stained with alamarBlue 72 hrs post-treatment. Data are means +/− SD of 3 independent experiments. n.s: non-significant, ***p < 0.001, two-tailed Student’s t-test. (I-J) HeLa cells synchronized as in Table S1B were transfected with indicated siRNAs and treated with MMC (1 μM) (I) or Gö6976 and MMC (1 μM each) (J) and harvested at indicated time points post-MMC. FANCI IPs were analyzed by western blot. (K) HeLa cells synchronized as in Table S1B were transfected with indicated siRNAs, treated with or without Gö6976 and MMC (1 μM each) and harvested 9 hrs post-MMC. FANCI IPs were analyzed by western blot. (L) HeLa cells transfected with indicated siRNAs were treated with or without Gö6976 (1 μM) and MMC (0.1 μM). Alamarblue was added 72 hrs post-treatment. Data are means +/− SEM of 3 independent experiments. **p < 0.01 ***p < 0.001, two-tailed Student’s t-test. (M-N) Schematic diagrams summarizing the results in panels D-I (M) and J-L (N). |
|
Monoubiquitination status impacts binding partner selection by FANCI.
(A) MDA-MB-231 and SAS cells treated with indicated doses of MMC and harvested at 9 hrs post-treatment. FANCI IPs were analyzed by western blot. Ub, monoubiquitinated form. (B-D) HeLa cells synchronized as in Table S1B were transfected with indicated siRNAs and treated with MMC (1 μM), harvested at 7.5 hrs post-treatment, immunoprecipitated with monoclonal PIDD1 (B-C) and FANCI (D) antibodies and analyzed by western blot. (E) FANCI–/– HCT116 cells transfected with sip53 were synchronized and reconstituted with indicated Flag-FANCI cDNAs as in Table S1C, treated with MMC (1 μM) and harvested 9 hrs later. FANCI IPs were analyzed by western blot. Cleaved C3 (cl.C3) and PARP (cl.) were assessed in 24 hr lysates. (F-K’’’) FANCI–/– HCT116 cells stably expressing shp53 were grown on cover slips, synchronized, reconstituted with indicated Flag-FANCI cDNAs, treated with or without MMC (1 μM) and harvested 19 hrs post-MMC. Cells were stained with DAPI (blue, F-K), anti-γH2A.X (red, F’-K’) and anti-active caspase-3 (green, F’’-K’’) and imaged by confocal microscopy (0.6 μm sections). Scale bar, 50 μm. (L) Quantification of γH2A.X and/or active caspase-3 positive cells, as shown in (F-K’’’), over three independent experiments. Data expressed as means +/−SD, ***p < 0.001, two-tailed Student’s t-test. (M) Model for the regulation of FANCI switch function by K523 monoubiquitination. |
Reprinted from Developmental Cell, 56, Shah, R.B., Kernan, J.L., van Hoogstraten, A., Ando, K., Li, Y., Belcher, A.L., Mininger, I., Bussenault, A.M., Raman, R., Ramanagoudr-Bhojappa, R., Huang, T.T., D'Andrea, A.D., Chandrasekharappa, S.C., Aggarwal, A.K., Thompson, R., Sidi, S., FANCI functions as a repair/apoptosis switch in response to DNA crosslinks, 2207-2222.e7, Copyright (2021) with permission from Elsevier. Full text @ Dev. Cell