- Title
-
Bicc1 and Dicer regulate left-right patterning through post-transcriptional control of the Nodal inhibitor Dand5
- Authors
- Maerker, M., Getwan, M., Dowdle, M.E., McSheene, J.C., Gonzalez, V., Pelliccia, J.L., Hamilton, D.S., Yartseva, V., Vejnar, C., Tingler, M., Minegishi, K., Vick, P., Giraldez, A.J., Hamada, H., Burdine, R.D., Sheets, M.D., Blum, M., Schweickert, A.
- Source
- Full text @ Nat. Commun.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
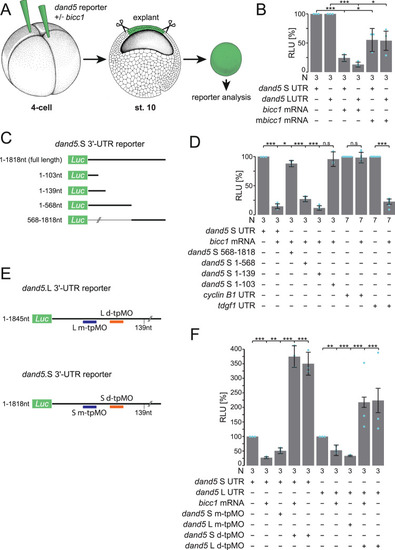
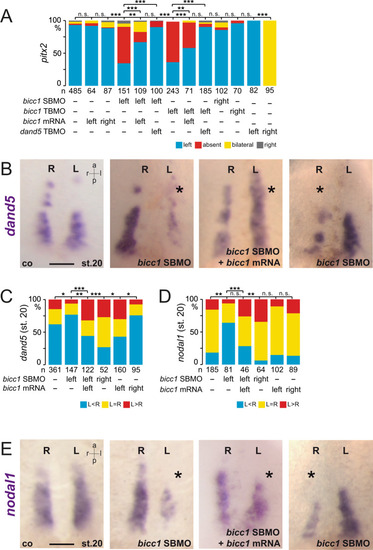
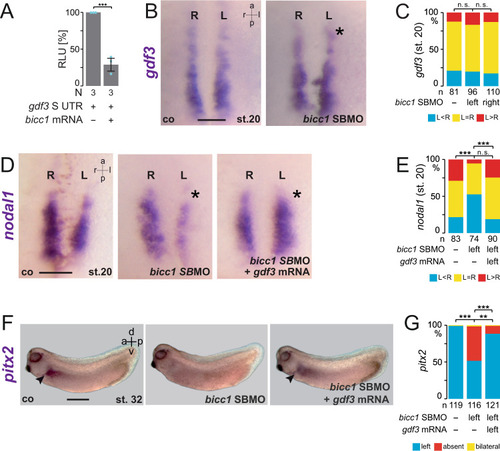
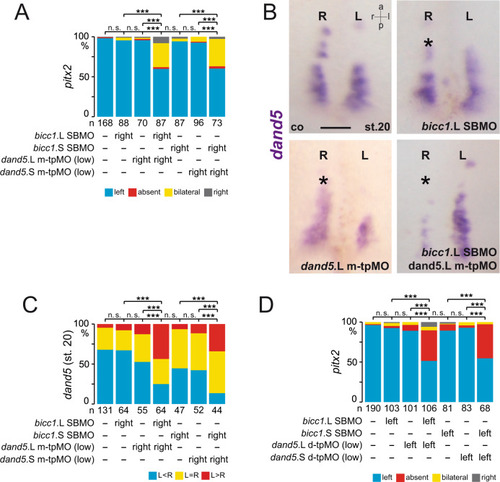
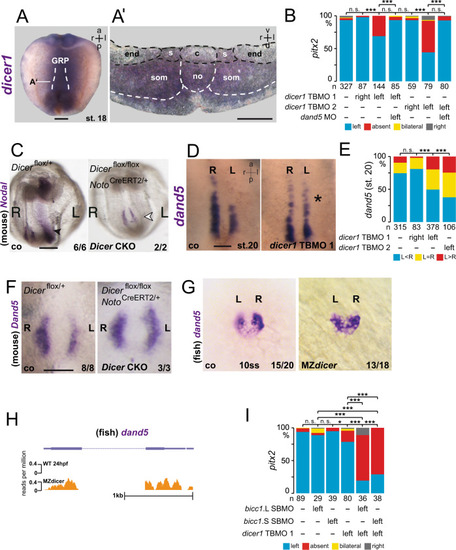
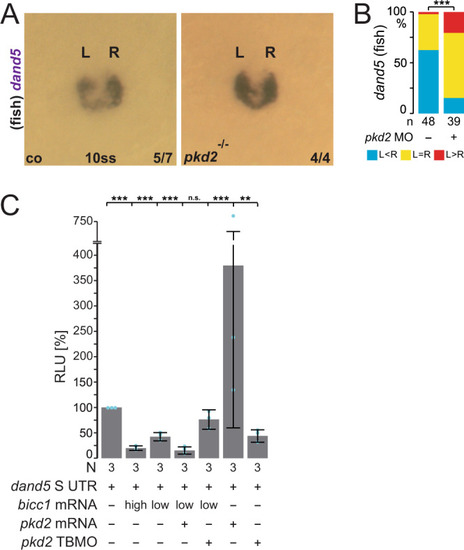
In the early neurula pre-flow stages, Bicc1 has two functions. Bicc1 assures |