- Title
-
A Dietary Cholesterol-Based Intestinal Inflammation Assay for Improving Drug-Discovery on Inflammatory Bowel Diseases
- Authors
- Silva, N.V., Carregosa, D., Gonçalves, C., Vieira, O.V., Nunes Dos Santos, C., Jacinto, A., Crespo, C.L.
- Source
- Full text @ Front Cell Dev Biol
|
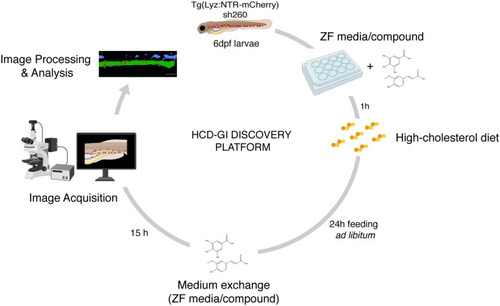
The high-cholesterol diet gut inflammation (HCD-GI) discovery platform. Schematic diagram illustrating the screening strategy for compounds modulating inflammation. Individual Tg(Lyz:NTR-mCherry)sh260 larvae, with fluorescent reporter expression in neutrophils, are distributed in 12-well plates with E3 zebrafish (ZF) medium supplemented with the compounds at 25 or 100 μM. After 1 h pre-treatment, larvae are fed with high-cholesterol diet (HCD) for 24 h. Larvae are then transferred to fresh medium supplemented with compounds were are kept for 15 h to allow inflammation and proper intestine emptying to occur. Larvae are finally fixed and imaged. Screening data are processed, and the neutrophilic inflammation index analyzed. Figure performed with images from Biorender. |
|
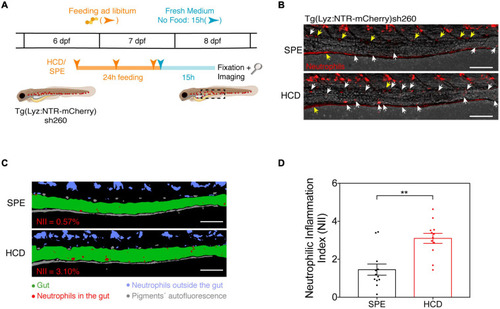
The HCD-gut inflammation (HCD-GI) assay. |
|
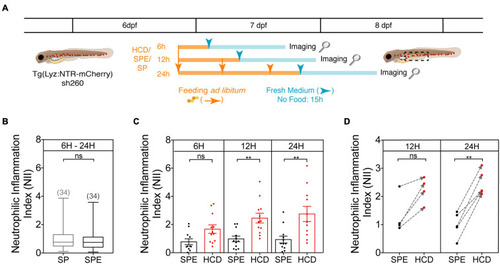
Protocol optimization for the HCD-GI assay. |
|
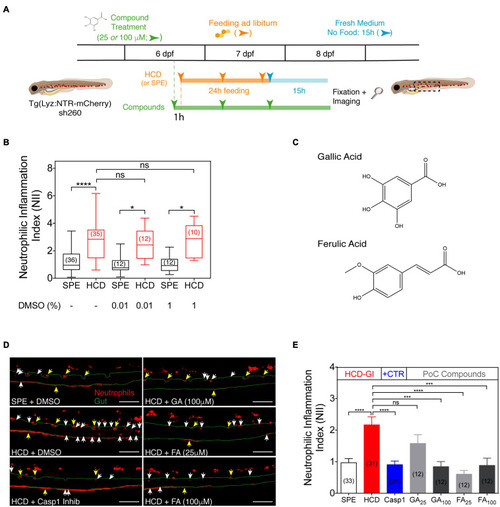
The HCD-GI discovery platform reveals an anti-inflammatory action of gallic acid (GA) and ferulic acid (FA) on zebrafish intestinal neutrophilic inflammation. |
|
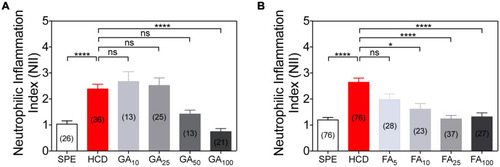
Dose-response anti-inflammatory action of gallic acid (GA) and ferulic acid (FA) using the HCD-GI assay. |
|
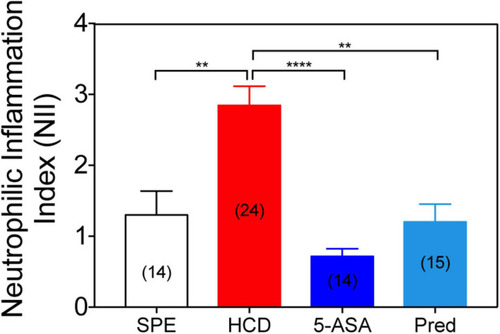
Treatment with conventional IBD therapeutics reduces the neutrophilic inflammation index. Neutrophilic inflammation index (NII) analysis in prednisolone (25 μM), mesalamine (5-ASA; 0.33 mM) or 1% DMSO-treated larvae. Data were aggregated from 2 (HCD) or 1 (SPE, Prednisolone, and 5-ASA) experimental replicates and pools of |