- Title
-
Insights into structure and dynamics of extracellular domain of Toll-like receptor 5 in Cirrhinus mrigala (mrigala): A molecular dynamics simulation approach
- Authors
- Rout, A.K., Acharya, V., Maharana, D., Dehury, B., Udgata, S.R., Jena, R., Behera, B., Parida, P.K., Behera, B.K.
- Source
- Full text @ PLoS One
|
The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was displayed. Evolutionary analyses were conducted on MEGA 6.0. |
|
( |
|
( |
|
( |
|
Structural imposed views of the top two cluster representatives (cluster 1: Green and cluster 2: Blue) obtained from clustering analysis using the snapshots from last 100 ns trajectory (A) and the evolution of secondary structure elements of |
|
( |
|
Per residue decomposition analysis displaying the contribution of each residue towards binding free energy in DrTLR5-Flagellin and CmTLR5-Flagellin complex systems. |
|
( |
|
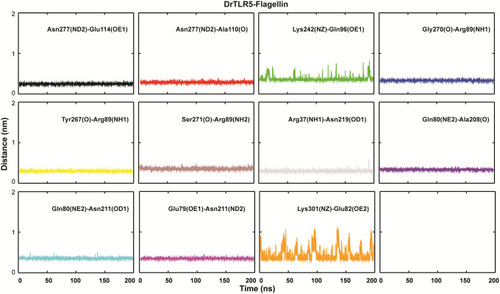
Inter-atomic distance profile of the important hydrogen bond forming amino acid pairs in |
|
Inter-atomic distance profile of the important hydrogen bond forming amino acid pairs in |