- Title
-
Ion channel profiling of the Lymnaea stagnalis ganglia via transcriptome analysis
- Authors
- Dong, N., Bandura, J., Zhang, Z., Wang, Y., Labadie, K., Noel, B., Davison, A., Koene, J.M., Sun, H.S., Coutellec, M.A., Feng, Z.P.
- Source
- Full text @ BMC Genomics
|
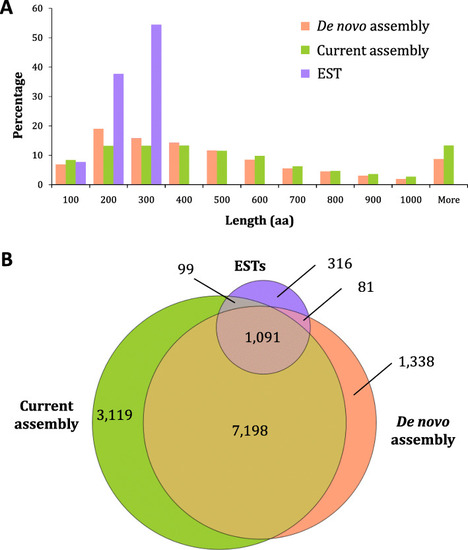
Workflow of quality control, assembly and prediction of protein-coding transcripts in the |
|
Comparison of predicted |
|
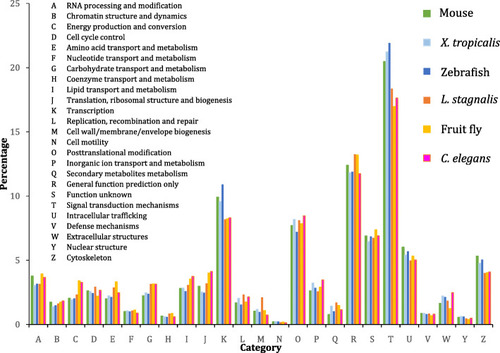
Comparison of KOG annotations of protein-coding transcripts expressed in the CNS of key vertebrate and invertebrate neuroscience model organisms (E-value <1E-5). Markedly, compared to invertebrates, vertebrate model organisms display increased percentage of transcripts involved in transcription, intracellular trafficking, and cytoskeleton. Meanwhile, compared to vertebrates, invertebrate model organisms display increased percentage of transcripts involved in energy production and conversion, amino acid transport and metabolism, carbohydrate transport and metabolism, and lipid transport and metabolism |
|
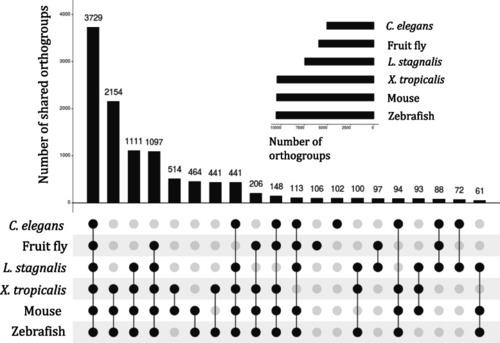
Membership of species in the orthogroups identified amongst protein-coding transcripts expressed in the CNS of |
|
Enriched GO terms of genes encoded by transcripts in the 3729 orthogroups shared amongst |
|
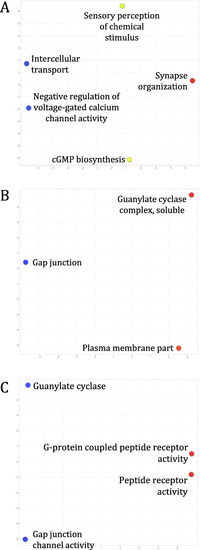
Enriched GO terms of genes encoded by transcripts in the 2154 orthogroups shared amongst only |
|
Enriched GO terms of genes encoded by transcripts in the 88 orthogroups shared amongst only |
|
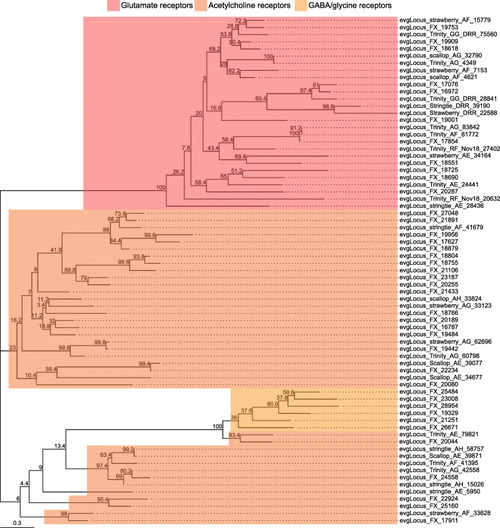
Ionotropic neurotransmitter receptor families identified in the |
|
K+ channel subtypes and families identified in the |
|
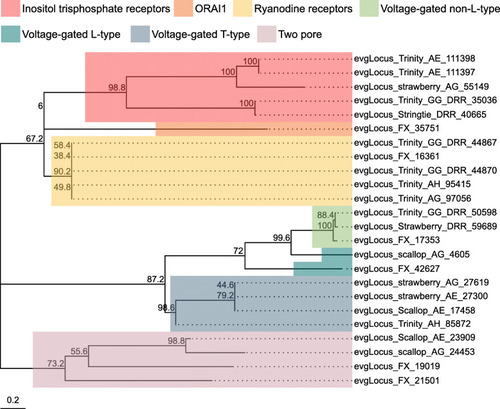
Ca2+ channel subtypes and families identified in the |
|
Na+ channel subtypes and families identified in the |
|
Cl− channel subtypes and families identified in the |
|
Cation channel subtypes and families identified in the |
|
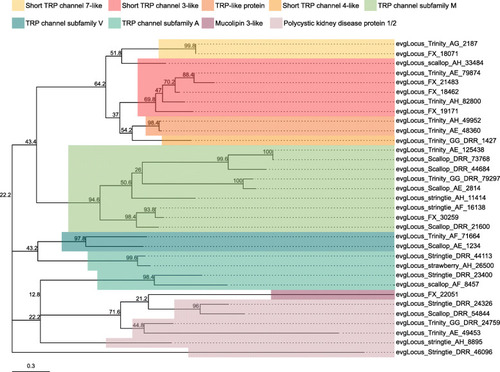
Transient receptor potential (TRP) channel subtypes and families identified in the |
|
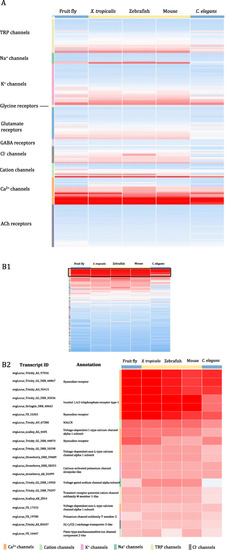
Heat maps of protein sequence similarity as measured by BLASTP bitscore between putative |