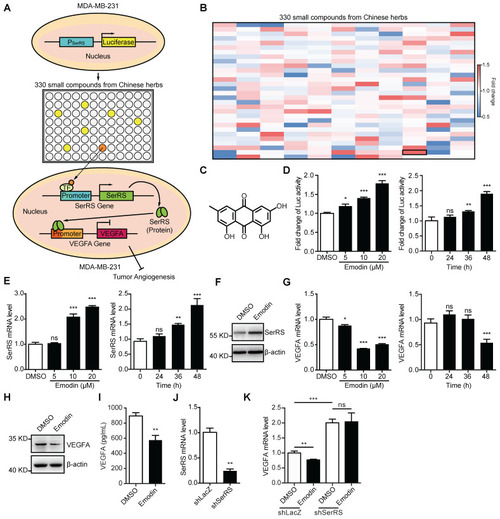
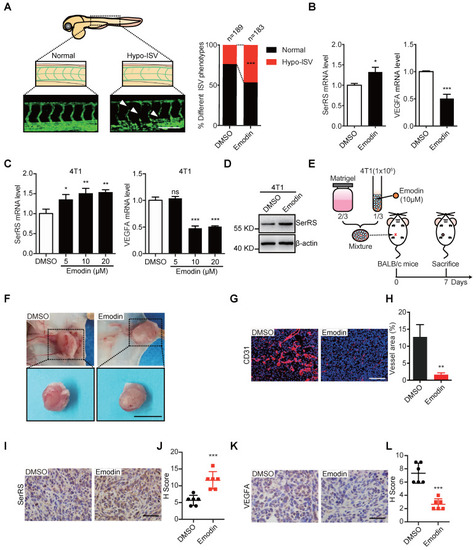
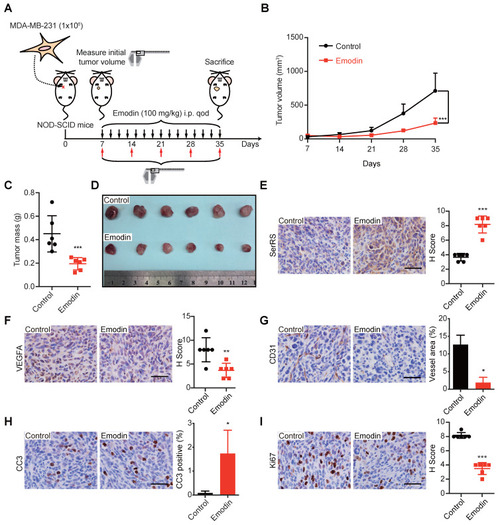
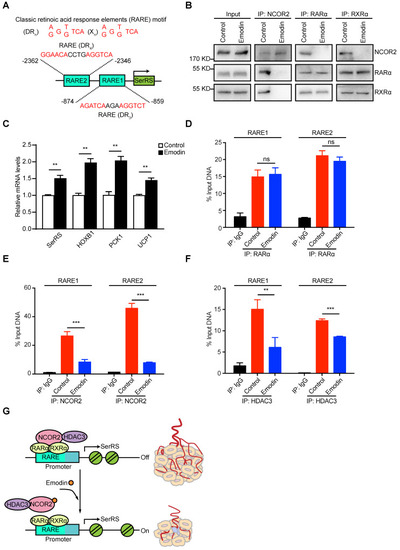
- Title
-
Herb-sourced emodin inhibits angiogenesis of breast cancer by targeting VEGFA transcription
- Authors
- Zou, G., Zhang, X., Wang, L., Li, X., Xie, T., Zhao, J., Yan, J., Wang, L., Ye, H., Jiao, S., Xiang, R., Shi, Y.
- Source
- Full text @ Theranostics
|
|
|
|
|
|
|
|
|
|
|
|