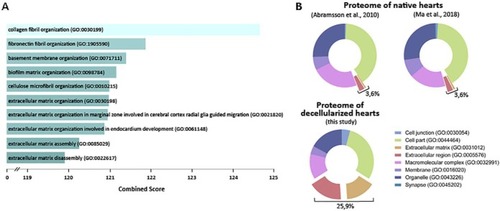
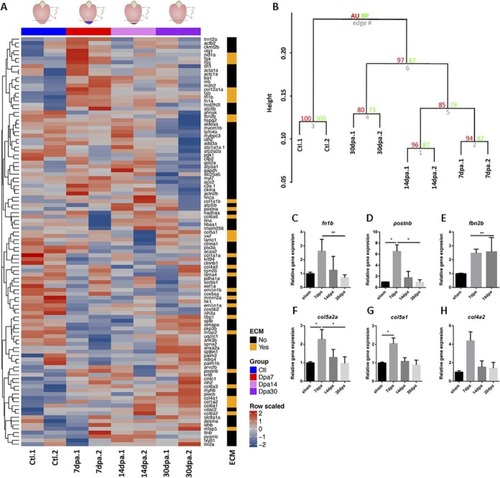
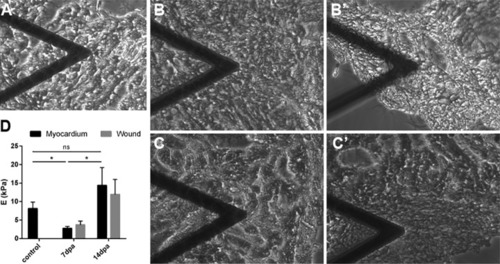
Decelularization protocol for zebrafish hearts.A, Representation of the protocol used to decellularize zebrafish ventricles. B, Images of the ventricles at different points of the decellularization protocol. From left to right, native ventricle, after 0.5% SDS, and at the end of the decellularization ptotocol (scale bar 500 μm). C–E, The efficiency of the decellularization process was characterized by DAPI staining (scale bar 50 μm) (C), spectrophotometric quantification of DNA (n = 3) (D), and Masson's trichrome (E, upper images) and Hematoxylin and Eosin (E, lower images) staining (scale bar 100 μm). F, Box plot representing an overall distribution of the average intensities of each protein in each sample group. ECM proteins and intracellular proteins have been independently plotted to better visualize the effect of decellularization in each class of proteins. G, The profile of specific ECM, cytoplasmic and nuclear proteins is plotted, showing loss of intracellular proteins and enrichment of ECM proteins during the decellularization protocol. Statistical significance of DNA content was analyzed with unpaired Student's t test. *, p < 0.05.
|