- Title
-
A rapid and effective method for screening, sequencing and reporter verification of engineered frameshift mutations in zebrafish
- Authors
- Prykhozhij, S.V., Steele, S.L., Razaghi, B., Berman, J.N.
- Source
- Full text @ Dis. Model. Mech.

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|
|
Expression of mutation reporters for chd7, hace1 and pycr1a mutations quantifies their frameshift efficiency. (A) Representative images of 16-18 hpf (∼16-somite stage) zebrafish embryos injected with the indicated mutation reporter mRNA from constructs made as described in Fig. 3 or uninjected. Fluorescence images of superfolder GFP (sfGFP) and TagRFP and merged images are shown. (B) Areas in the somite regions of these embryos were quantified by measuring fluorescence intensities of both sfGFP and TagRFP, adjusted for background in the corresponding channel, and the ratio of these intensities was calculated to account for injection variability and used for plotting the data and statistical analysis. For chd7 mutation reporters, the difference between wild-type (n=15) and 2-bp deletion mutant (n=12) mutation reporters was large and very significant (P=2.034e−15). Embryos injected with hace1 wild-type (n=10) and 8-bp insertion mutant (n=12) cDNA fragments did not show any significant difference. The pycr1a wild-type (n=41) and 71-bp deletion mutant (n=28) reporters had a small but significant difference (P=7.776e−13). ***P<0.001. The injections were performed twice on different days with very similar results and embryos from one of the injections were subjected to imaging and statistical analysis of fluorescent protein intensities ratios. Student's two-tailed t-test was performed for each experiment. PHENOTYPE:
|
|
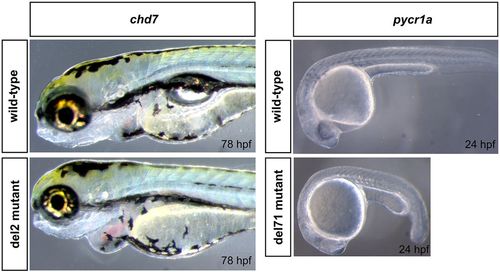
Phenotypes of chd7 and pycr1a mutants. The phenotype of chd7 del2 homozygous mutants was most clearly observed after 72 hpf and included smaller eyes, enlarged heart with edema and failure to inflate the swimbladder; images show fish at the 78 hpf stage as an example. For pycr1a del71 mutants, the observed abnormality was a pronounced developmental delay at 24 hpf, as shown in the image, that persists up to about 48 hpf and disappears afterwards. |
|
Mutation reporters show that an artificial hace1 4-bp deletion in the middle of hace1 cDNA fragment induces efficient frameshifting. A: Representative images of 16-18 hpf (ca 16-somite) stage zebrafish embryos injected with the wild-type hace1 mutation reporter or a reporter construct with a 4-bp deletion mutation after the first 106 codons. Fluorescence images of superfolder GFP (sfGFP), TagRFP and a merged image are shown. B: Areas in the somite regions of these embryos were quantified by measuring fluorescence intensities of both sfGFP and TagRFP, adjusted for background in the corresponding channel and the ratio of these intensities was calculated to account for injection variability and used for plotting the data and statistical analysis. The difference between wild-type (n = 17) and 4-bp deletion mutant (n = 18) mutation reporters was large and very significant (Student’s twotailed t-test: p-value = 3.27e-14). The fact of p-value being less than 0.001 is marked with “***”. The embryos from two independent injections were subjected to imaging and statistical analysis of fluorescent protein intensities ratios. |