- Title
-
Advances in the Study of Heart Development and Disease Using Zebrafish
- Authors
- Brown, D.R., Samsa, L.A., Qian, L., Liu, J.
- Source
- Full text @ J Cardiovasc Dev Dis
|
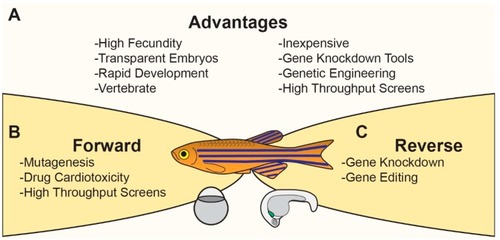
Zebrafish Model System. Schematic illustrating ( |
|
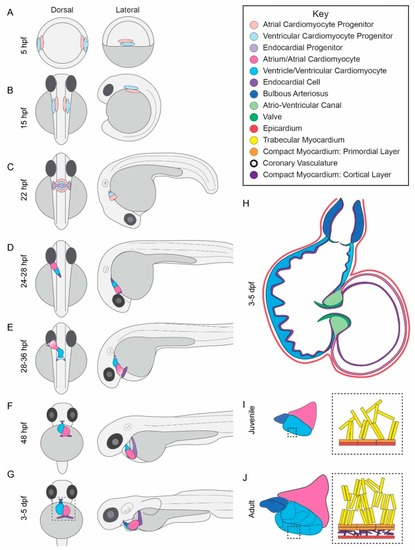
Zebrafish heart development. ( |
|
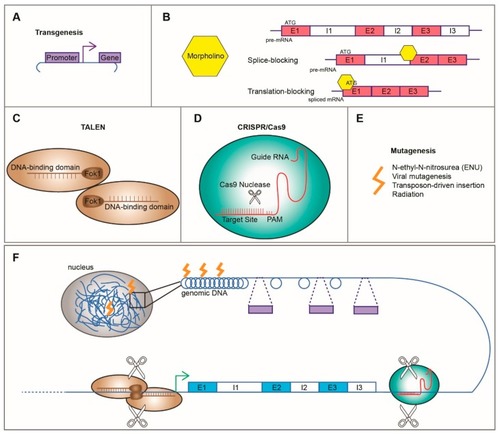
Common tools used in zebrafish include ( |