- Title
-
The homologous recombination component EEPD1 is required for genome stability in response to developmental stress of vertebrate embryogenesis
- Authors
- Chun, C., Wu, Y., Lee, S.H., Williamson, E.A., Reinert, B.L., Jaiswal, A.S., Nickoloff, J.A., Hromas, R.A.
- Source
- Full text @ Cell Cycle
|
EEPD1 deletion causes developmental delay and death during Zebrafish embryogenesis. (A) Depletion of the HR nuclease EEPD1 by MO injection into zygotes results in delayed embryonic development or death. Averages ±SD for two determinations are plotted. A total of 115-240 embryos were scored per condition. *** indicates P < 0.0001 by Fisher exact test for the combined data from the two determinations. (B) Morphologic abnormalities in developing zebrafish depleted of EEPD1, especially in the somite region; representative embryos are shown at 2, 5, and 6 days post-fertilization (dpf). |
|
EEPD1 depletion prevents replication stress signaling. (A) Representative confocal immunofluorescent photomicrographs of the somite region of zebrafish embryos stained for γ-H2Ax. (B) Percentage of somite nuclei >5 γ-H2Ax foci. Values are averages (±SD) for 3-6 embryos per condition, 56–375 nuclei scored per embryo. *** indicates P < 0.0001, t-tests. |
|
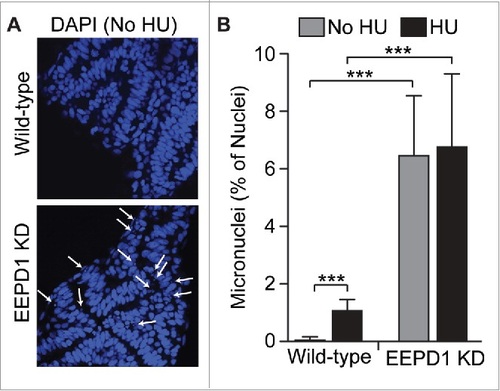
EEPD1 depletion in zebrafish embryos causes genome instability with or exogenous replication stress induced by 4 hr treatment with 5 mM HU. (A) Representative photomicrographs of somite nuclei in zebrafish embryos stained with DAPI. Arrows indicate micronuclei. (B) Percentage of somite nuclei displaying micronuclei. Values are averages (±SD) for 6-16 embryos per condition, 115-421 nuclei scored per embryo. *** indicates P ≤ 0.0001, t-tests. PHENOTYPE:
|