- Title
-
In vivo analysis of Hyaloid vasculature morphogenesis in zebrafish: A role for the lens in maturation and maintenance of the Hyaloid
- Authors
- Hartsock, A., Lee, C., Arnold, V., Gross, J.M.
- Source
- Full text @ Dev. Biol.
|
Stage I: arrival of fli1a:GFP+ cells at the ventral eye. (A) Schematic of 1-2 dpf imaging paradigm and maximum projection data. Drawings not to scale. (B) fli1a:GFP+ cells (arrowheads) arrive at the ventral eye between 18 and 20 hpf. Precursor cells proceed towards the lens through the choroid fissure. Lens position indicated by dashed blue line, retina by dashed beige line. hh:mm. Scale bar=50 µm D: Dorsal, V: Ventral, N: Nasal, T: Temporal, La: Lens anterior, Lp: Lens posterior. EXPRESSION / LABELING:
|
|
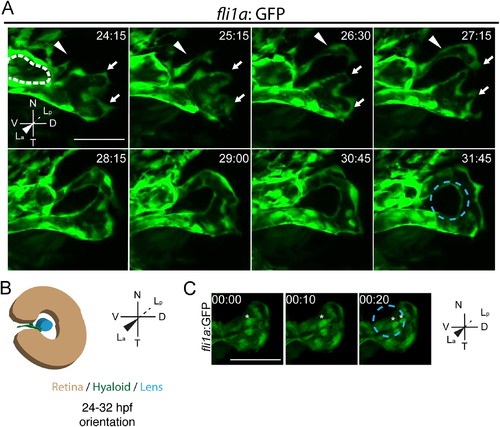
Stage I: hyaloid loop formation. (A) Still maximum projection images from time-lapse movies highlighting formation of the hyaloid loop. Nasal (white arrow) and temporal (white arrowhead) oriented sprout morphogenesis to form the hyaloid loop. Dashed white line outlines cross section of vessel from which the hyaloid loop sprouts. (B) Schematic of maximum projection data. (C) Stills from time-lapse movie showing mitotic cells in the vessel stalk (asterisks) during formation of hyaloid loop. Lens position in B and C indicated by dashed blue line. hh:mm. All scale bars=50 µm. D: Dorsal, V: Ventral, N: Nasal, T: Temporal, La: Lens anterior, Lp: Lens posterior. |
|
Stage II: formation of a branched hyaloid network. All images are still maximum projections from time-lapse movies. (A) Schematic of imaging angles and maximum projection data. Drawings not to scale. (B) Increases in vessel network complexity within the posterior hyaloid. Asterisk marks the nasally oriented sprout of the hyaloid loop. (C) Angiogenesis within the hyaloid increases vessel network complexity. Asterisk marks site of break in vessel, arrowheads mark sprouting event that does not form a new connection, arrows mark sprouting event that generates a new loop within the vessel network. (D) Connection of the hyaloid to the annular ring (arrowhead). Lens position in B and C indicated by dashed blue line. hh:mm. Scale bars=50 µm. D: Dorsal, V: Ventral, N: Nasal, T: Temporal, La: Lens anterior, Lp: Lens posterior. EXPRESSION / LABELING:
|
|
Stage III: refinement of the hyaloid network. (A) Schematic of imaging angles and maximum projections data. Drawings not to scale. (B) Maximum projections of 3-5 dpf hyaloid in transverse imaging view demonstrating the continued reduction and refinement of the vessel network. (C) Still images from time-lapse movies demonstrating the further reduction in vessel complexity as fibers connecting adjacent vessels retract (asterisk). hh:mm. (A,B) Scale bar=50µm; inset scale bar=5 µm. D: Dorsal, V: Ventral, N: Nasal, T: Temporal, La: Lens anterior, Lp: Lens posterior. EXPRESSION / LABELING:
|
|
Quantification of hyaloid growth. (A) DIC and GFP maximum projections images of dissected lenses from fli1a:GFP embryos at 12 h intervals from 2 to 5 dpf. Dashed line indicates location of the lens. Scale bars=50 µm. D: Dorsal, V: Ventral, La: Lens anterior, Lp: Lens posterior (B) Quantification of anterior progression of the hyaloid over time. (C) Reduction in hyaloid vessel branching over time. *p≤0.01, **p≤0.001, ***p≤1×10-10. n=number of samples analyzed. |
|
Microangiography demonstrates that the hyaloid vessel is fully enclosed by 5 dpf. (A) Maximum projection images of the eye of a fli1a:GFP (green) embryo injected with 50 kDa rhodamine dextran (red). Merged image highlights that rhodamine is detected throughout the vitreous. Iso-surface rendering of merged image. (B-D) Iso-surface renderings of maximum projections collected from the eyes of 3-5 dpf fli1a:GFP (green) embryos injected with 50 kDa rhodamine dextran (red) and imaged either dorsally or ventrally. Arrowhead highlights rhodamine containment in a fli1a:GFP vessel and arrows highlight rhodamine outside of vessels. hh:mm. Scale bars=50 µm. D: Dorsal, V: Ventral, N: Nasal, T: Temporal, La: Lens anterior, Lp: Lens posterior. |
|
mab21l2au10 mutants possess defects in lens formation. (A) Images of phenotypically wild-type sibling and mab21l2au10 mutants at 4 dpf. High-magnification views of the eyes of mild and severe mab21l2au10 mutants. (B) Transverse cryossections of wild-type and severe mab21l2au10 mutants at 1 and 4 dpf highlighting the lack of a lens in severe mab21l2au10 mutants. Scale bars=50 µm. (C) Genomic sequences from wild-type, heterozygous and mab21l2au10 mutants. mab21l2au10 mutants possess an A->T transversion at position 301, resulting in a premature stop codon at amino acid 101. (D) Schematic of protein length of wild-type and mab21l2au10 mutant. (E) High magnification view of eye in sibling (wild-type) and mab21l2au10 mutant embryo injected with mab21l2-GFP (rescue). (F) Quantification of lens phenotype after mab21l2-GFP injection. |
|
The lens is required for Stages II and III hyaloid maturation and maintenance. All images are stills from time-lapse movies from severe mab21l2au10; fli1a:GFP mutants. (A) Hyaloid precursor cell recruitment is delayed by ~2 h in mab21l2au10 mutants but recruitment is otherwise normal. (B) Hyaloid loop formation occurs in the absence of a lens. (C) Stage II formation of a branched hyaloid network is disrupted in mab21l2au10 mutants. Hyaloid cells appear disorganized and dynamic, having not coalesced into obvious vessels. (D) The hyaloid in mab21l2au10 mutants still makes contact anteriorly with the annular vessel. (E) Microangiography demonstrates that at 3d pf, the mab21l2au10 mutant hyaloid is not enclosed anteriorly, and rhodamine (red) fills the retina. hh:mm. Beige dashed line in A, B and D indicate outlines of the retina. All scale bars=50 µm. D: Dorsal, V: Ventral, N: Nasal, T: Temporal, La: Lens anterior, Lp: Lens posterior. EXPRESSION / LABELING:
PHENOTYPE:
|
Reprinted from Developmental Biology, 394(2), Hartsock, A., Lee, C., Arnold, V., Gross, J.M., In vivo analysis of Hyaloid vasculature morphogenesis in zebrafish: A role for the lens in maturation and maintenance of the Hyaloid, 327-39, Copyright (2014) with permission from Elsevier. Full text @ Dev. Biol.