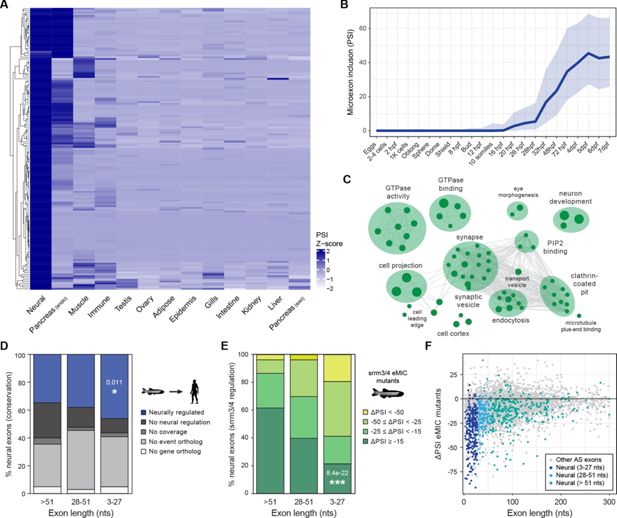
Fig. 1 Neural microexon program in zebrafish. (A) Heatmap showing relative inclusion levels of neural microexons identified in zebrafish (Supplementary file 1). The value for each tissue type corresponds to the average inclusion in the samples for that tissue in VastDB (Supplementary file 1). Only events with sufficient read coverage in >10 tissue groups are plotted (N=157) and missing values were imputed. (B) Inclusion of neural microexons along embryo development (egg to 7 days post-fertilization [dpf]). Thick line corresponds to the median PSI and the shades to the first and third quartile of the PSI distribution. (C) Enriched Gene Ontology (GO) categories among genes harboring neural microexons in zebrafish. GO terms were grouped into networks by ClueGO. (D) Evolutionary conservation of zebrafish neural exons of different length group at the genomic and tissue-regulatory level compared with human. Exons conserved at the regulatory level (blue) are those with enriched inclusion in neural samples (ΔPSI ≥15) also in human. Those with no neural regulation (black) are conserved at the genomic level (Irimia et al., 2009), but are not neurally enriched. Those conserved but with insufficient coverage to assess regulation are indicated as "No coverage" (dark grey). P-value corresponds to a two-sided Fisher’s Exact test for neural conservation vs. others between exons >51 nts and 3–27 nts. (E) Distribution of exons of different length by the level of srrm3/4 misregulation in larva or retina (see Methods). P-value corresponds to a two-sided Fisher’s Exact test for non-regulated exons (ΔPSI ≥ –15) vs. others between exons >51 nts and 3–27 nts. (F) Change in inclusion levels [ΔPSI (enhancer of microexons domain (eMIC) mutant-WT)] for all exons shorter than 300 bp. Dots with different blue colors correspond to neural exons of different length.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife