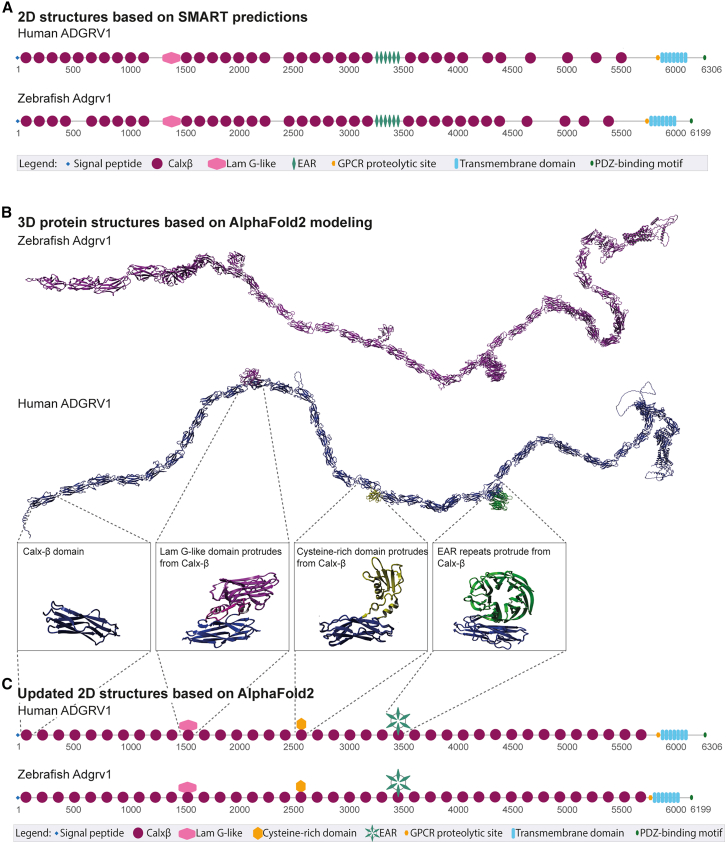
Figure 1
(A) Schematic representation of the protein domain structure of the large isoform (isoform B) of human and zebrafish ADGRV1, based on 2D-SMART predictions. Both proteins exhibit a repetitive domain architecture, comprising a signal peptide, 35 Ca2+-binding calcium exchanger β domains (Calxβ), a laminin G-like domain (LamG-like), 6 epilepsy-associated repeats (EAR), a G-protein-coupled receptor (GPCR) proteolytic site, a seven-transmembrane region, and an intracellular region with a C-terminal class I PDZ-binding motif. Numbers indicate amino acid positions. (B)