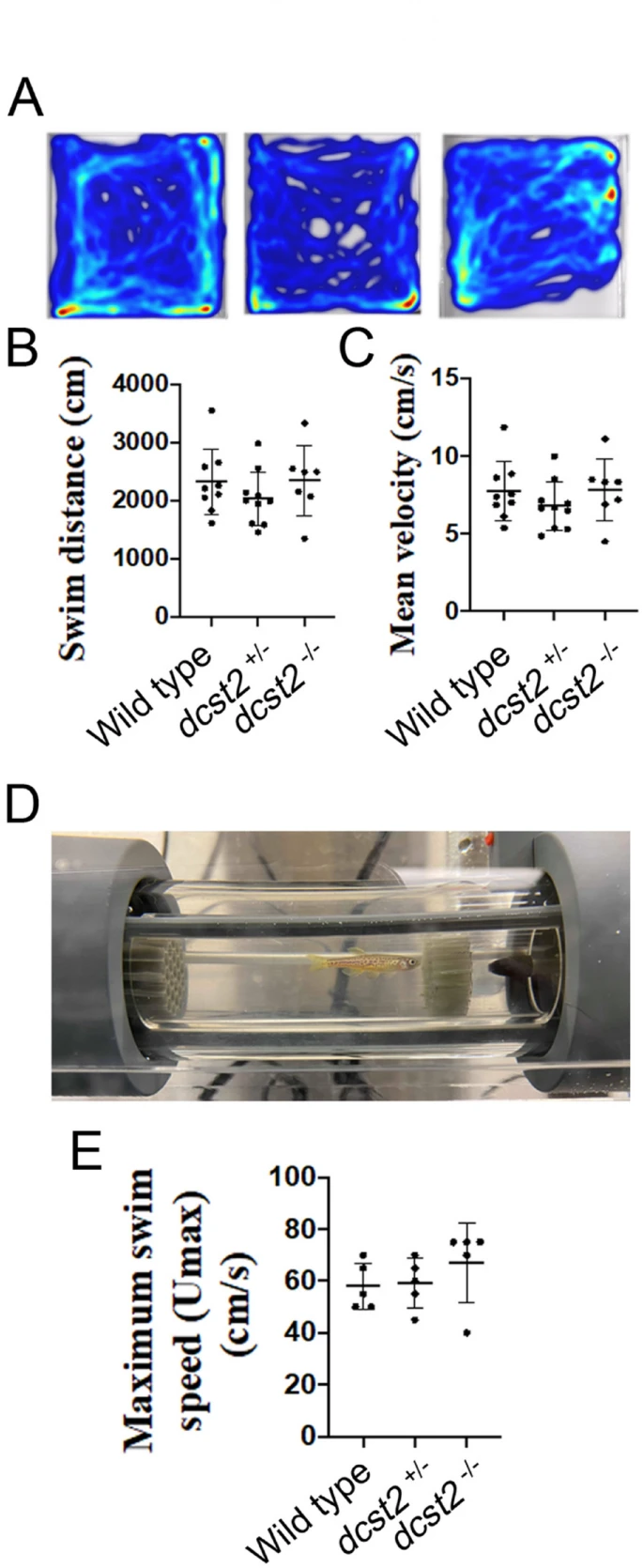
Fig. 1 Genomic structure of zebrafish dcst2 and the CRISPR gRNA target site used to generate a knockout mutant. Zebrafish dcst2 contains 16 exons and is located on chromosome sixteen. Blue exons denote translated exons and white exons are the Untranslated Regions (UTR). A frame shift mutation in exon 3 was identified in a founder mutant line resulting in premature stop codons. Yellow highlighted nucleotides indicate the PAM sequence and red highlighted nucleotides indicate the guide RNA target sequence. Genotypic ratios generated from crosses between two dcst2+/− mutant zebrafish depicted as a pie chart (B). Transcriptional expression levels of dcst2 relative to actin in the testis confirmed that our mutation likely is being degraded by nonsense-mediated decay (C). Asterisks represent a statistically significant difference (p < 0.05). D, A comparison between muscle and testis dcst2 expression indicating that dcts2 is not expressed in muscle. Asterisks represent a statistically significant difference (p < 0.05). E, Tabulation of male body weight of each fish from each genotype. Asterisks represent a statistically significant difference (p < 0.05). However, this finding was slightly underpowered with the power of the performed test being 0.701 with alpha (α) value set at 0.05
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Mol. Genet. Genomics