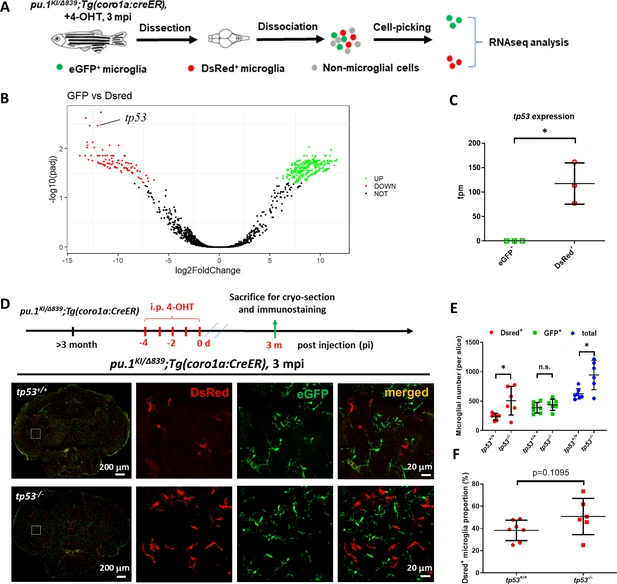
Fig. 5 Inactivation of Tp53 largely restored the number of pu.1-deficient microglia in mosaic condition. (A) The experimental setup for the isolation of eGFP+ and DsRed+ microglia from pu.1KI/Δ839;Tg(coro1a:CreER) adult brain at 3 mpi for transcriptomic analysis. (B) The volcano plot of differentially expressed genes (DEG) between eGFP+ and DsRed+ microglia at 3 mpi. (C) Relative expression of tp53 in eGFP+ (n=3) and DsRed+ (n=3) microglia at 3 mpi by transcripts per million (TPM). (D) The experimental setup for pu.1 conditional knockout in wild-type and tp53-/- adult zebrafish, and the representative images of midbrain cross section of pu.1KI/Δ839;Tg(coro1a:CreER) and pu.1KI/Δ839;tp53-/-;Tg(coro1a:CreER) fish at 3 mpi. (E) Quantification of the number of DsRed+, eGFP+, and total (DsRed + eGFP) microglia in pu.1KI/Δ839;Tg(coro1a:CreER) (n=7) and pu.1KI/Δ839;tp53-/-;Tg(coro1a:CreER) (n=6) fish at 3 mpi. (F) Quantification of the proportion of DsRed+ microglia in pu.1KI/Δ839;Tg(coro1a:CreER) (n=7) and pu.1KI/Δ839;tp53-/-;Tg(coro1a:CreER) (n=6) fish at 90 dpi. n.s.=not significant, p>0.05; *p<0.05.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife