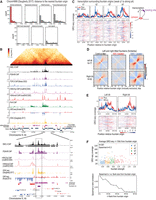
Fig. 7 Fountain origins are close to active enhancers. (A) Histogram of genomic distance between identified fountain origins in this paper and the annotated regions of ChromHMM in (Daugherty et al. 2017). Active enhancers are highlighted in red. (B) Genome browser snapshot of Hi-C and other transcription related features such as topoisomerase-II, H3K27ac, H3K4me3, Pol II, GRO-seq, and ATAC-seq (top). Zoom-in near a fountain origin is shown at the bottom. (C) Average profile of GRO-seq data from Kruesi et al. (2013) split into plus (red) and minus (blue) strands is plotted with respect to the fountain origins. The fountains are grouped into four quantiles based on their strength. On the right, the cartoon highlights that origins of strong fountains are often close to upstream of transcription. (D) Left- and right-directed fountains were identified using the fontanka tool (Galitsyna et al. 2023). The fountain origins of the two groups are mutually exclusive. (E) GRO-seq plot centered around the left- and right-directed fontanka fountain origins. (F) Scatterplot showing relationship between fountain strength of the fountain origin and the signal mean of GRO-seq (Kruesi et al. 2013) in 20 kb region centered at the fountain origin (top). Sweep of Spearman's correlation as a function of flanking region used to average GRO-seq signal (bottom).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Genome Res.