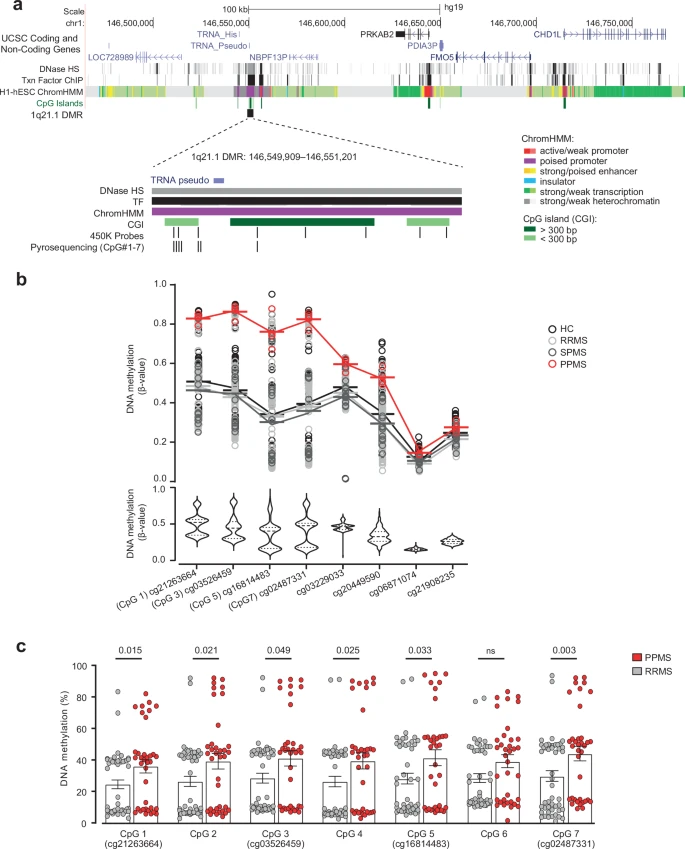
Fig. 2 Hypermethylation of the 1q21.1 locus in primary progressive multiple sclerosis patients. a Genomic annotation of the identified differentially methylated region (DMR) in 1q21.1, including 450K and pyrosequenced CpG probes and regulatory features, i.e. CpG island (CGI) and chromatin state segmentation by hidden Markov model (ChromHMM) from ENCODE/Broad. b Dot plot and violin plot of DNA methylation differences at the identified DMR on chromosome 1: 146549909–146551201 (hg19) in PPMS (n = 4) compared to RRMS (n = 120), SPMS (n = 16), and HC (n = 139), obtained using Illumina 450K in cohort 1. c Replication of methylation (mean ± SEM) at 7 CpGs (CpG 1–7, including four 450 K probes) in an independent cohort (nRRMS = 48, nPPMS = 36, non-parametric two-sided Mann–Whitney U test) using pyrosequencing. MS multiple sclerosis, PPMS primary progressive MS, RRMS relapsing-remitting MS, SPMS secondary progressive MS, HC healthy controls, BOMS bout-onset MS, hiPSC human induced pluripotent stem cells, DNase HS DNase I hypersensitive sites, Txn factor ChIP transcription factor chromatin immunoprecipitation. Source data are provided as a Source Data file.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Nat. Commun.