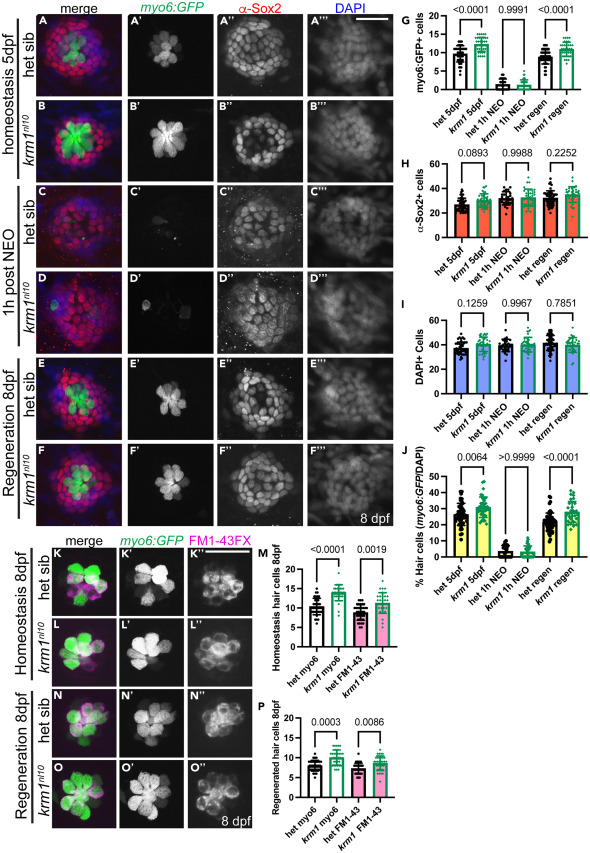
Fig. 2 krm1nl10 larvae form supernumerary hair cells following regeneration in posterior lateral line neuromasts (A–B‴) Confocal projections of neuromasts in heterozygous sibling and krm1nl10 larvae with Tg(myosin6b:GFP)w186 expression in hair cells (green), α-Sox2 antibody-labeled support cells (red) and DAPI-labeled nuclei (blue) at 5dpf NM prior to exposure to NEO. (C–D‴) Heterozygous sibling and krm1nl10 mutant at 1 h post NEO exposure. (E–F‴) Heterozygous sibling and krm1nl10 mutant 3 days post NEO exposure. (G–J) Quantification of Tg(myosin6b:GFP)w186 positive hair cells, α-Sox2 antibody-labeled support cells at 5dpf pre-NEO exposure, and the percentage of hair cells Tg(myosin6b:GFP)w186 -positive cells/DAPI-labeled cells) at 1-h post NEO exposure, and at 8dpf, 3 days post NEO exposure.). 5dpf: het sibling n = 48 NMs (9 fish), krm1nl10 n = 38 NM (9 fish); 1h-post NEO: het sibling n = 25 NMs (7 fish), krm1nl10 n = 36 NM (7 fish); 8dpf: het sibling n = 57 NMs (11 fish), krm1nl10 n = 34 NM (10 fish). (K–L″) Confocal projections of live images showing FM1-43 incorporation (magenta) in Tg(myosin6b:GFP)w186-positive hair cells (green) at 8dpf in heterozygous sibling krm1nl10 mutant NMs under homeostatic conditions. (M) Quantification of labeled hair cells during homeostasis, heterozygous sibling n = 36 NM (9 fish), krm1nl10 n = 31 NM (10 fish). (N–O″) FM1-43 incorporation (magenta) in Tg(myosin6b:GFP)w186-positive hair cells (green) at 8dpf heterozygous sibling and krm1nl10 mutant NMs under regeneration conditions. (P) Quantification of regenerated hair cells, heterozygous sibling n = 34 NM (10 fish), krm1nl10 n = 33 NM (10 fish). All data presented as mean ± SD, Kruskal-Wallis test, Dunn’s multiple comparisons test. Scale bar = 20 μm.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ iScience