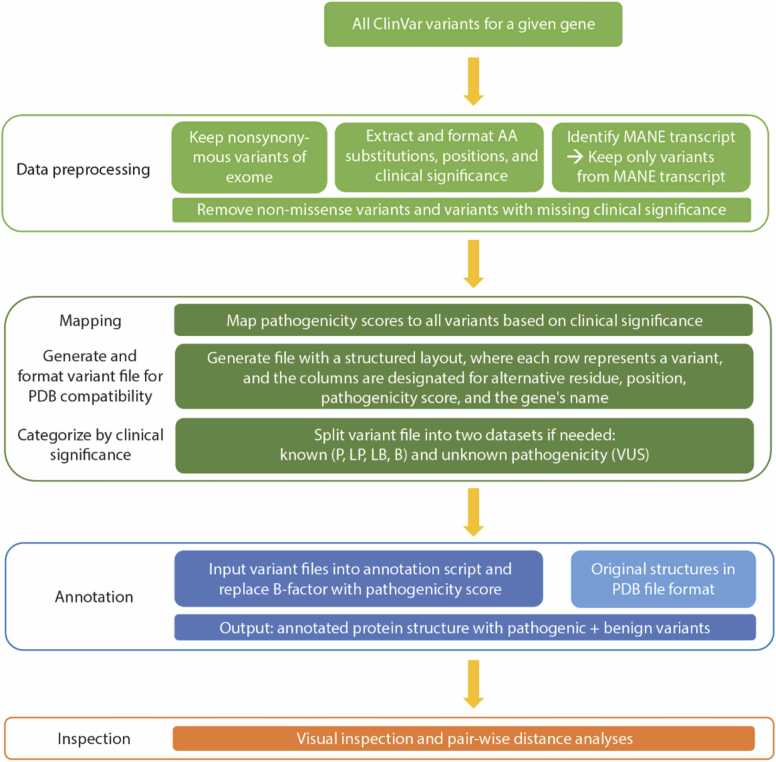
Fig. 1
Overview of the workflow for variant annotation of protein structures. All ClinVar variants must first undergo data preprocessing and parsing steps to ensure that only nonsynonymous variants within the relevant transcript that also have data about clinical significance move on to the next steps. The clinical significance categories (P, LP, VUS, LB, B) are assigned a pathogenicity score (see Methods), which is then used to map variants onto the protein structure for further visualization, inspection, and analysis. The other ClinVar class of variants with “conflicting interpretations” is omitted from analysis.