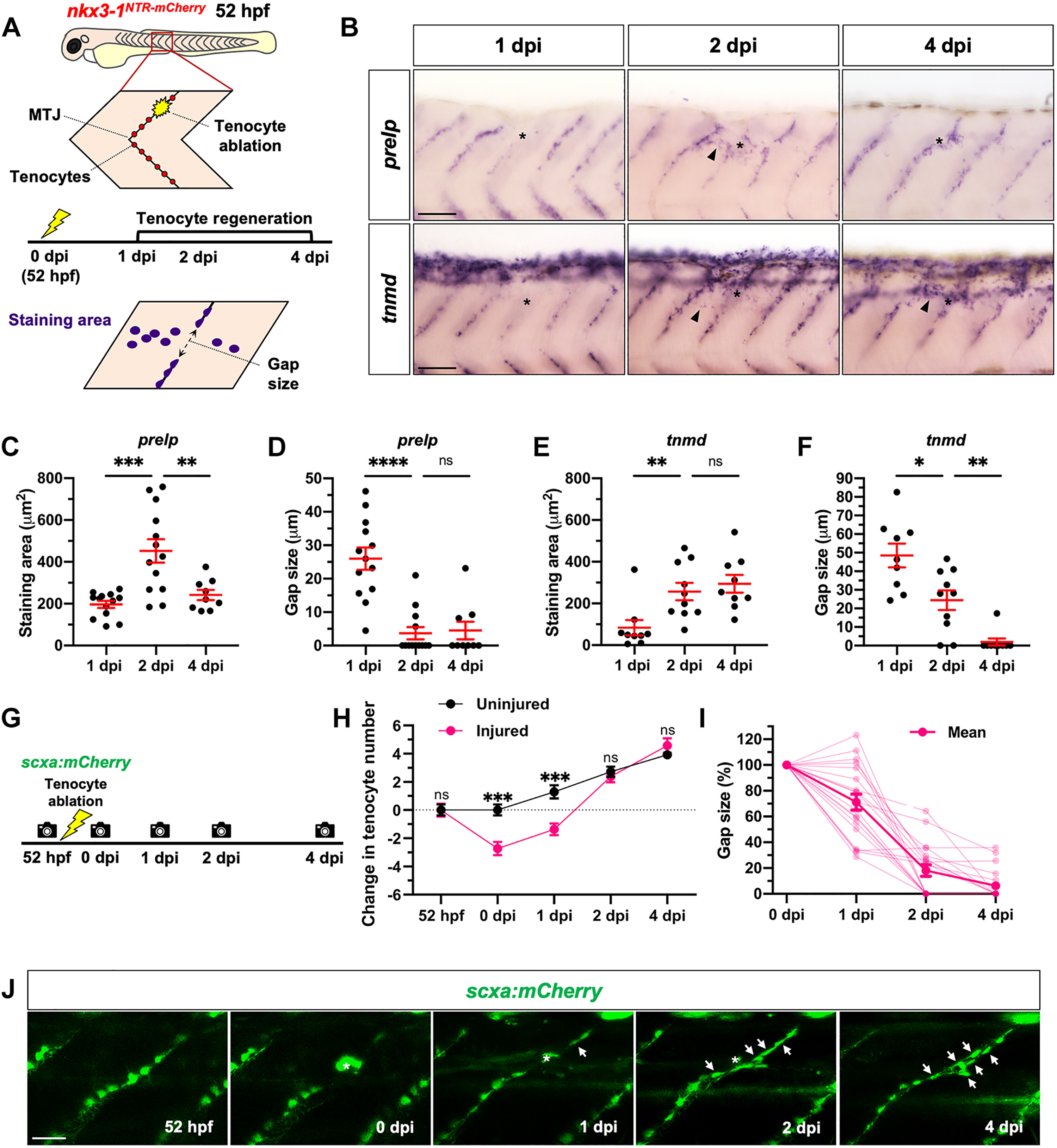
Fig. 2 Zebrafish can regenerate trunk tenocytes after laser ablation. (A) Schematic representation of tenocyte ablation protocol and quantification shown in (B) to (F). (B) In situ hybridization for tenocyte markers, prelp (top) and tnmd (bottom), at 1, 2, and 4 dpi showing increased gene expression (arrowheads) around the injury region (asterisks) at 2 dpi. n = 9 to 13 embryos per probe at each stage. (C to F) Quantification of staining area and gap size in stained embryos as described in (A). (G) Experimental protocol to track tenocyte regeneration in live scxa:mCherry embryos. (H) Change in tenocyte number from 52 hpf to 4 dpi along injured and uninjured MTJs. (I) Quantification of gap between adjacent tenocytes at the injury site in each embryo from 0 to 4 dpi. (J) Representative images of one injured embryo from 52 hpf to 4 dpi, with several newly regenerated tenocytes (arrows) arising at the injured MTJ. Autofluorescent scar from laser ablation indicated by asterisks. n = 14 uninjured and 19 injured MTJs from 19 embryos. All data shown as mean ± SEM. Statistics: Mann-Whitney U test [(C) to (F)]; Sidak’s multiple comparisons (H). Significance: ns (not significant), P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Scale bars, 50 μm (B) and 25 μm (J).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Sci Adv